Chapter 9 General Analysis
- In this page, users can explore the most genomics matrix datasets of UCSC Xena repository with common analysis methods.
- As Chapter 8.3 describes, the initial step involves the selection of interesting datasets from Repository page.
- In the following, we will take the “TCGA-BLCA.htseq_fpkm.tsv” dataset as an example to demonstrate how to perform various analyses.
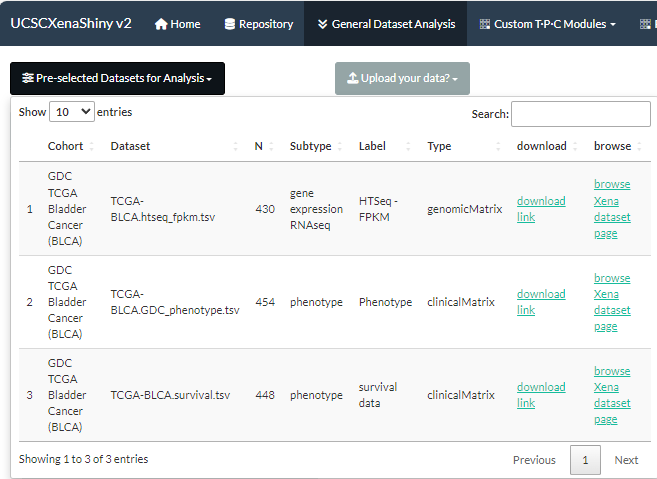
Figure 9.1: The selection of TCGA-BLCA.htseq_fpkm.tsv dataset
9.1 Scatter-Correlation
Compute and visualize the correlation between two molecules based on vis_identifier_cor() function.
- Firstly, select two molecules from candidate dataset(s);
- Click the “Submit” button to perform analysis and the visualization result along with raw data will be display in the middle.
- Finally, the plot can be saved with size and format options.
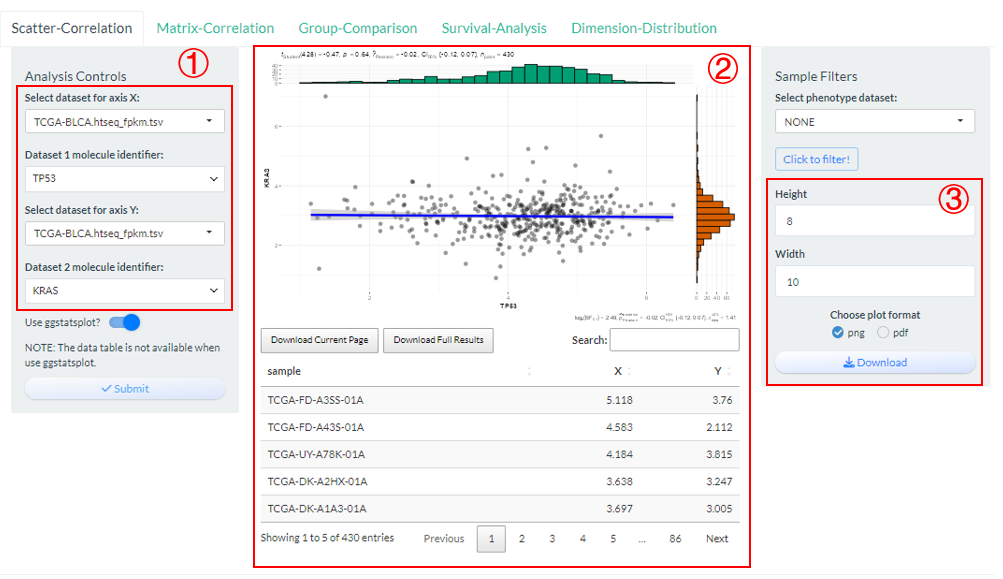
Figure 9.2: The steps of Scatter-Correlation analysis
Notably, two molecules can come from different datasets, but there must be intersecting samples.
9.2 Matrix-Correlation
Compute and visualize the pair-wise correlation among multiple molecules based on vis_identifier_multi_cor() function.
- Firstly, select multiple molecules from one candidate dataset;
- Modify several analysis and visualization parameters;
- Click the “Submit” button to perform analysis and the visualization result along with analyzed data will be display in the middle.
- Finally, the plot can be saved with size and format options.
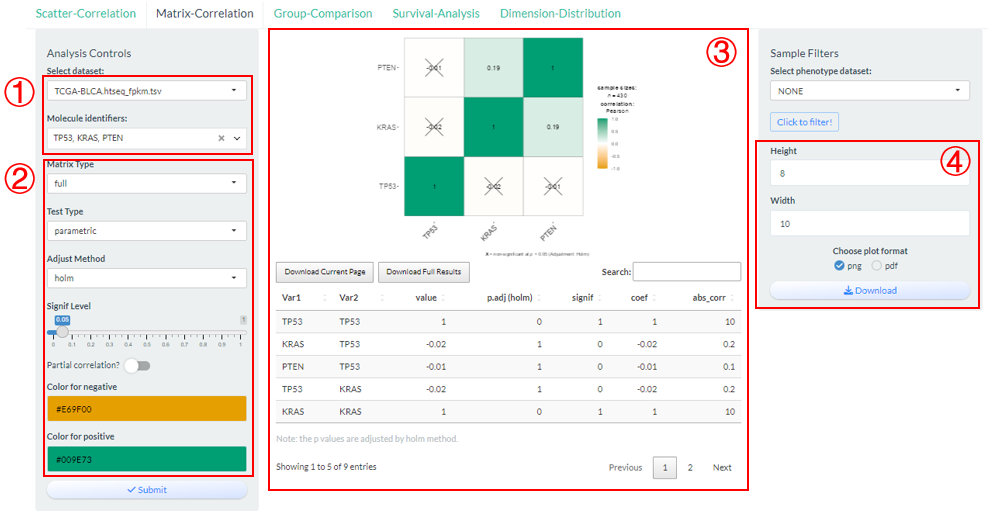
Figure 9.3: The steps of Matrix-Correlation analysis
9.3 Group-Comparison
Compare the differences in the distribution of values for one molecule under a specified phenotypic grouping based on vis_identifier_grp_comparison() function.
- Select a dataset and use one of its columns as the basis for grouping. (dataset 1)
- Select a dataset and use one of its numeric columns as the values to compare. (dataset 2)
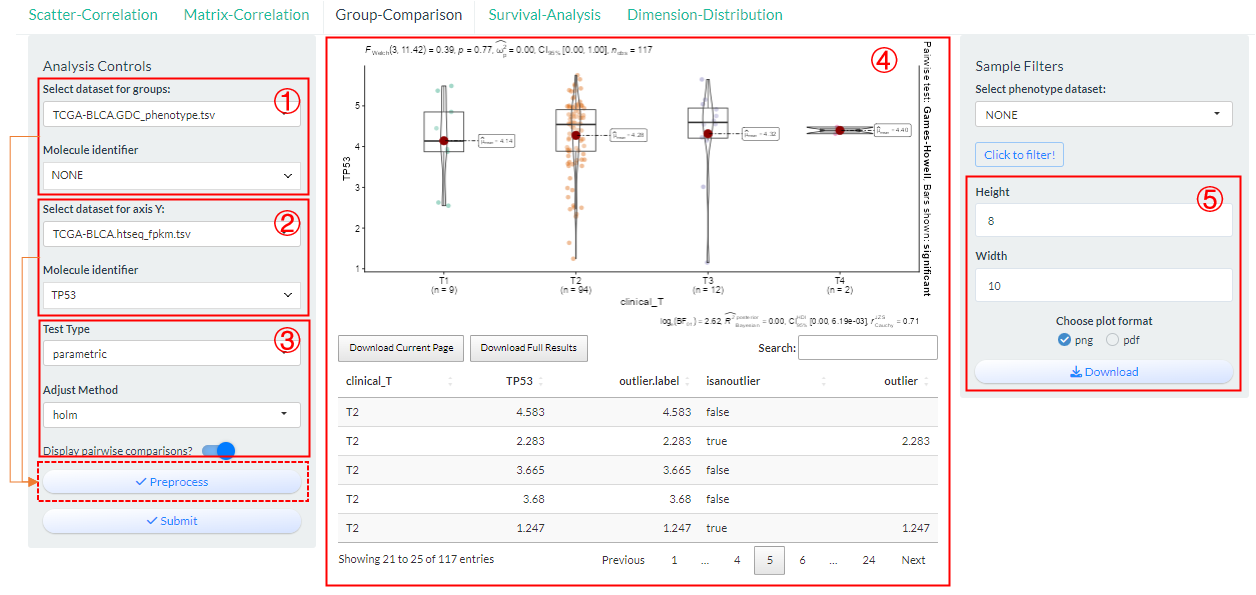
Figure 9.4: The steps of Group-Comparison analysis
Based on above two selections, then set sample groups by clicking “Preprocess” button.
- Step1: From the columns of dataset 1, select the sample id column (
Sample column), which is usually left as a default. Then, select a column (Group column) as the basis for grouping. If it is a numeric column, you can set the percentile cutoffs by the slider widget.- Step2: From the columns of dataset 2, select the sample id column (
Sample column) and molecule column (Value column).- Step3: Click the “Click to joint the data” button to finally generate grouping result. If there are no problems, exit this interface.
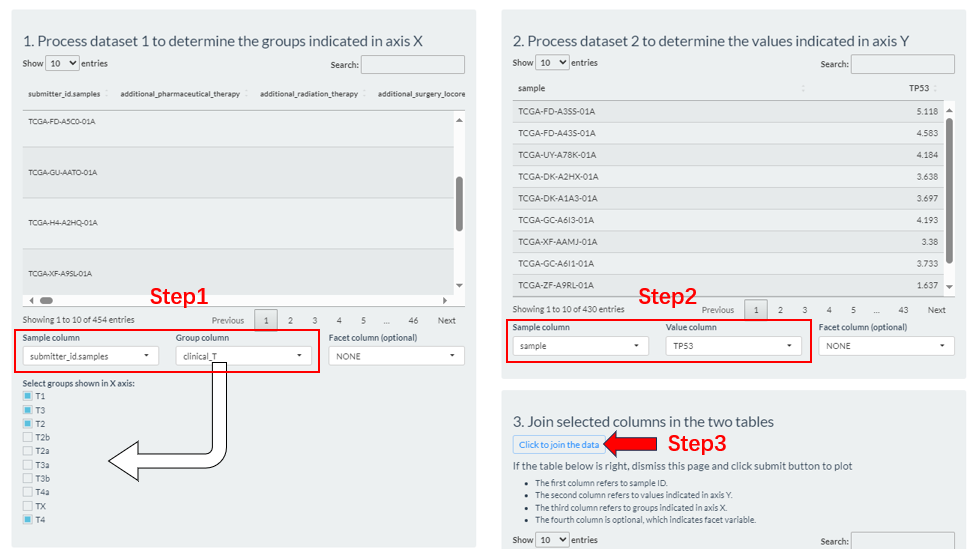
Figure 9.5: The Group-Comparison grouping via the Preprocess button
- Modify several analysis parameters;
- Click the “Submit” button to perform analysis and the visualization result along with raw data will be display in the middle.
- Finally, the plot can be saved with size and format options.
9.4 Survival-Analysis
Perform the grouping log-rank survival analysis, especially for TCGA related datasets based on vis_identifier_grp_surv() function.
- Select a dataset and use one of its columns as the basis for grouping. (dataset 1)
- Select one survival dataset with event time and status columns. (dataset 2)
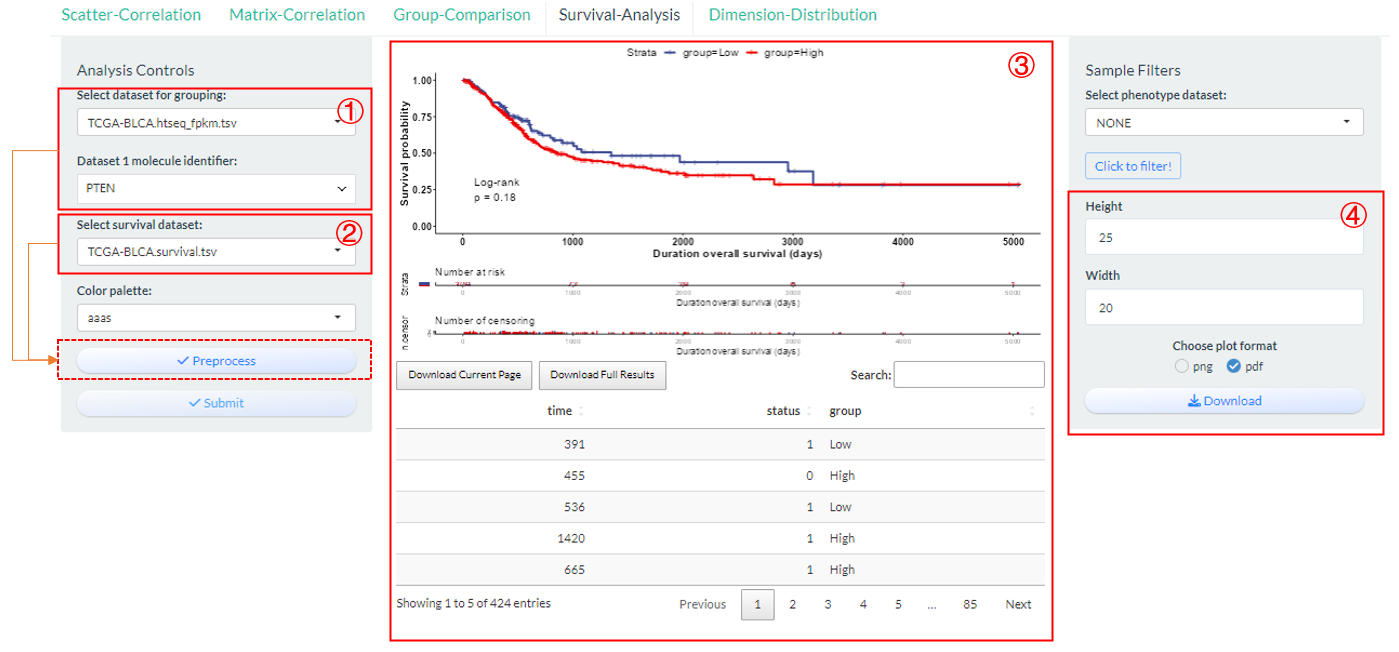
Figure 9.6: The steps of Survival-Analysis
Based on above two selections, then set sample groups by clicking “Preprocess” button.
- Step1: From the columns of dataset 1, select the sample id column (
Sample column), which is usually left as a default. Then, select a column (Group column) as the basis for grouping. If it is a numeric column, you can set the percentile cutoffs by the slider widget or automatically calculate its best cutoff.- Step2: From the columns of dataset 2, select the sample id column (
Sample column) and two survival related column (Time column,Status column).- Step3: Click the “Click to joint the data” button to finally generate grouping result. If there are no problems, exit this interface.
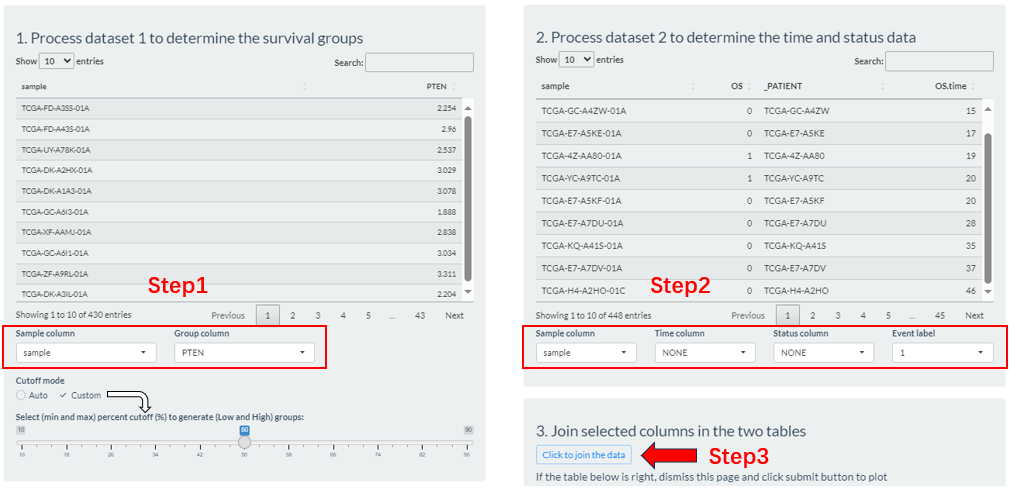
Figure 9.7: The Survival-Analysis grouping via the Preprocess button
- Click the “Submit” button to perform analysis and the visualization result along with raw data will be display in the middle.
- Finally, the plot can be saved with size and format options.
9.5 Dimension-Distribution
Perform the dimension reduction analysis based on multiple molecules of one genomics matrix datasets based on vis_identifier_dim_dist() function.
- Select a dataset and use one of its columns as the basis for grouping. (dataset 1)
- Select a genomics matrix dataset and further multiple molecular columns via three ways. (dataset 2).
- “Select”: One-by-one selection;
- “Pathway”: batch selection under one pathway;
- “File”: Upload of identifier file.
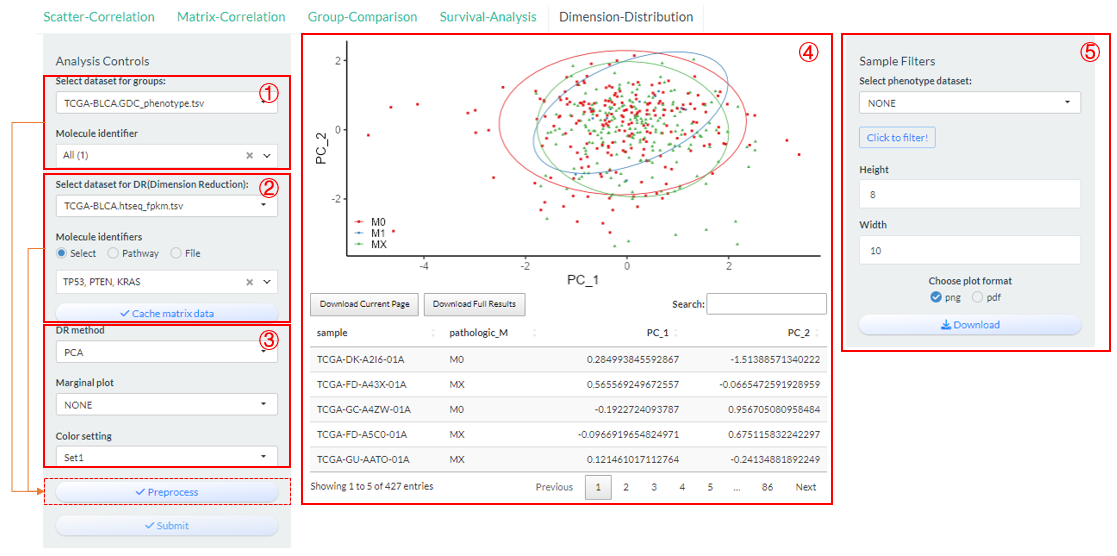
Figure 9.8: The steps of Dimension-Distribution
Based on above two selections, then set sample groups by clicking “Preprocess” button.
- Step1: From the columns of dataset 1, select the sample id column (
Sample column), which is usually left as a default. Then, select a column (Group column) as the basis for grouping. If it is a numeric column, you can set the percentile cutoffs by the slider widget.- Step2: From the columns of dataset 2, select the sample id column (
Sample column) and molecule columns (Value column).- Step3: Click the “Click to joint the data” button to finally generate grouping result. If there are no problems, exit this interface.
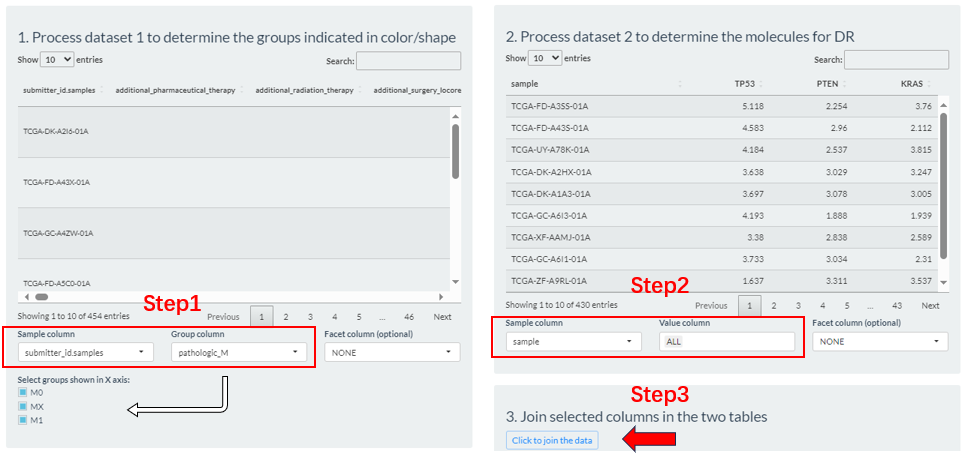
Figure 9.9: The Dimension-Distribution grouping via the Preprocess button
- Modify several analysis and visualization parameters;
- three DR methods (PCA/UMAP/TSNE) are supported.
- Click the “Submit” button to perform analysis and the visualization result along with analyzed data will be display in the middle.
- Finally, the plot can be saved with size and format options.