Chapter 3 Built-in Datasets
We also have curated tens of tumor non-omics annotation datasets with supplementary features from UCSC Xena repository or other resources for extensive analysis. Notably, datasets with large amounts of data are uploaded to Zenodo. They can be loaded via load_data() function.
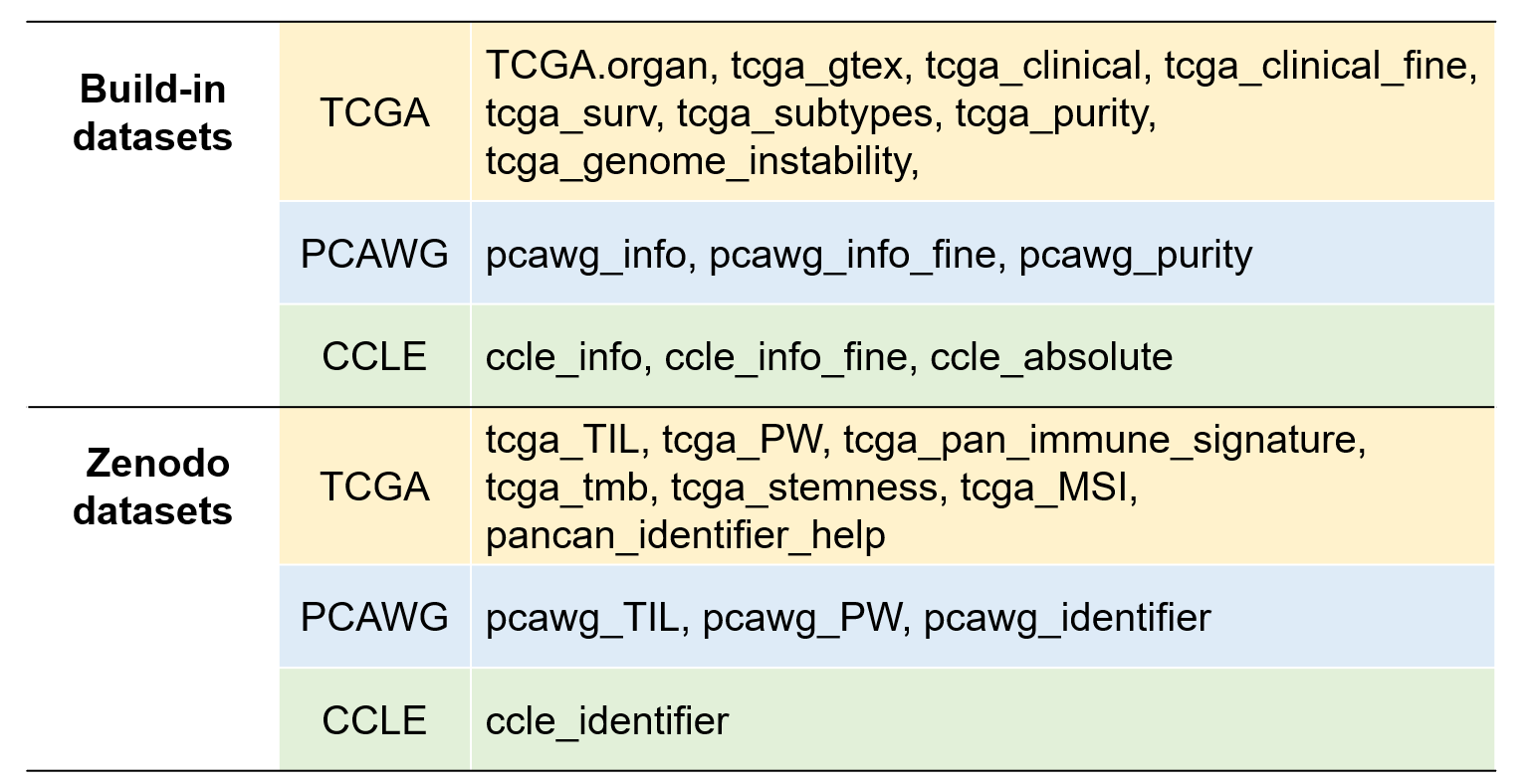
Figure 3.1: UCSCXenaShiny built-in datasets
3.1 TCGA
TCGA.organ: Detailed information of 33 TCGA projects
## TCGA Detail organ
## 1 BRCA breast invasive carcinoma breast
## 2 PRAD prostate adenocarcinoma prostate
## 3 OV ovarian serous cystadenocarcinoma ovary
## 4 PCPG pheochromocytoma & paraganglioma
## 5 GBM glioblastoma multiforme brain
## 6 HNSC head & neck squamous cell carcinomatcga_gtex: Merged information of TCGA and GTEx samples
## sample tissue type type2
## 1 TCGA-D3-A1QA-07 SKCM SKCM_tumor_TCGA tumor
## 2 TCGA-DE-A4MD-06 THCA THCA_tumor_TCGA tumor
## 3 TCGA-J8-A3O2-06 THCA THCA_tumor_TCGA tumor
## 4 TCGA-J8-A3YH-06 THCA THCA_tumor_TCGA tumor
## 5 TCGA-EM-A2P1-06 THCA THCA_tumor_TCGA tumor
## 6 TCGA-J8-A4HW-06 THCA THCA_tumor_TCGA tumortcga_clinical: Common phenotypes of TCGA samples from Table S1 of the paper.tcga_clinical_fine: Basic phenotypes of TCGA samples
## # A tibble: 6 × 8
## Sample Cancer Age Code Gender Stage_ajcc Stage_clinical Grade
## <chr> <chr> <dbl> <chr> <chr> <chr> <chr> <chr>
## 1 TCGA-OR-A5J1-01 ACC 58 TP MALE Stage II <NA> <NA>
## 2 TCGA-OR-A5J2-01 ACC 44 TP FEMALE Stage IV <NA> <NA>
## 3 TCGA-OR-A5J3-01 ACC 23 TP FEMALE Stage III <NA> <NA>
## 4 TCGA-OR-A5J4-01 ACC 23 TP FEMALE Stage IV <NA> <NA>
## 5 TCGA-OR-A5J5-01 ACC 30 TP MALE Stage III <NA> <NA>
## 6 TCGA-OR-A5J6-01 ACC 29 TP FEMALE Stage II <NA> <NA>tcga_surv: Survival data of TCGA samples from Table S1 of the paper.
## sample OS OS.time DSS DSS.time DFI DFI.time PFI PFI.time
## 1 TCGA-OR-A5J1-01 1 1355 1 1355 1 754 1 754
## 2 TCGA-OR-A5J2-01 1 1677 1 1677 NA NA 1 289
## 3 TCGA-OR-A5J3-01 0 2091 0 2091 1 53 1 53
## 4 TCGA-OR-A5J5-01 1 365 1 365 NA NA 1 50
## 5 TCGA-OR-A5J6-01 0 2703 0 2703 0 2703 0 2703
## 6 TCGA-OR-A5J7-01 1 490 1 490 NA NA 1 162tcga_subtypes: Subtype information of TCGA samples from Pan-Cancer Atlas Hub
## sampleID Subtype_mRNA Subtype_DNAmeth Subtype_protein Subtype_miRNA
## 1 TCGA-02-0001-01 LGr4 LGm5 <NA> <NA>
## 2 TCGA-02-0003-01 LGr4 LGm5 K1 <NA>
## 3 TCGA-02-0004-01 LGr4 <NA> K1 <NA>
## 4 TCGA-02-0006-01 <NA> LGm5 <NA> <NA>
## 5 TCGA-02-0007-01 unclassified LGm4 <NA> <NA>
## 6 TCGA-02-0009-01 LGr4 LGm4 <NA> <NA>
## Subtype_CNA Subtype_Integrative Subtype_other Subtype_Selected
## 1 <NA> <NA> Mesenchymal-like GBM_LGG.Mesenchymal-like
## 2 <NA> <NA> Mesenchymal-like GBM_LGG.Mesenchymal-like
## 3 <NA> <NA> <NA> GBM_LGG.NA
## 4 <NA> <NA> Mesenchymal-like GBM_LGG.Mesenchymal-like
## 5 <NA> <NA> Classic-like GBM_LGG.Classic-like
## 6 <NA> <NA> Classic-like GBM_LGG.Classic-like
## Subtype_Immune_Model_Based
## 1 <NA>
## 2 <NA>
## 3 <NA>
## 4 <NA>
## 5 <NA>
## 6 <NA>tcga_purity: tumor purity related information of TCGA samples from Supplementary Data 1 of the paper.
## # A tibble: 6 × 7
## sample cancer_type ESTIMATE ABSOLUTE LUMP IHC CPE
## <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 TCGA-OR-A5J1-01 ACC 0.937 NaN 0.977 0.8 0.925
## 2 TCGA-OR-A5J2-01 ACC 0.918 NaN 0.617 0.95 0.898
## 3 TCGA-OR-A5J3-01 ACC 0.967 NaN 0.925 0.8 0.947
## 4 TCGA-OR-A5J4-01 ACC NaN NaN 0.920 0.8 0.866
## 5 TCGA-OR-A5J5-01 ACC 0.976 NaN 1 0.8 0.978
## 6 TCGA-OR-A5J6-01 ACC 0.874 NaN 0.744 0.88 0.840tcga_genome_instability: tumor genome instability related information of TCGA samples from here
## sample purity ploidy Genome_doublings Cancer_DNA_fraction
## 1 TCGA-02-0003-01 0.90 1.96 0 0.90
## 2 TCGA-02-0033-01 0.45 1.94 0 0.44
## 3 TCGA-02-0047-01 0.69 1.99 0 0.69
## 4 TCGA-02-0055-01 0.55 3.13 1 0.65
## 5 TCGA-02-2466-01 0.96 1.93 0 0.96
## 6 TCGA-02-2470-01 0.53 2.00 0 0.53
## Subclonal_genome_fraction
## 1 0.01
## 2 0.13
## 3 0.14
## 4 0.69
## 5 0.17
## 6 0.003.2 PCAWG
pcawg_info: Common phenotypes of TCGA samples from PCAWG Xena Hubpcawg_info_fine: Basic phenotypes of TCGA samples
## # A tibble: 6 × 5
## Sample Project Age Gender Type
## <chr> <chr> <dbl> <chr> <chr>
## 1 SP1003 BLCA-US 53 female tumor
## 2 SP1007 BLCA-US 53 female normal
## 3 SP10084 BRCA-US 64 female tumor
## 4 SP1009 BLCA-US 84 male tumor
## 5 SP10150 BRCA-US 48 female tumor
## 6 SP101515 OV-AU 54 female tumorpcawg_purity: tumor purity related information of PCAWG samples from PCAWG Xena Hub
## # A tibble: 6 × 6
## icgc_specimen_id purity ploidy purity_conf_mad wgd_status wgd_uncertain
## <chr> <dbl> <dbl> <dbl> <chr> <lgl>
## 1 SP101724 0.885 3.36 0.039 wgd FALSE
## 2 SP79365 0.774 2.00 0.022 no_wgd FALSE
## 3 SP98853 0.8 2.43 0.011 no_wgd FALSE
## 4 SP47708 0.837 1.83 0.03 no_wgd FALSE
## 5 SP106808 0.92 1.64 0.003 no_wgd FALSE
## 6 SP102816 0.596 1.97 0.006 no_wgd FALSE3.3 CCLE
ccle_info: Common phenotypes of CCLE samples from Broad Instituteccle_info_fine: Basic phenotypes of CCLE samples
## # A tibble: 6 × 5
## Sample Site_Primary Gender Histology Type
## <chr> <chr> <chr> <chr> <chr>
## 1 1321N1_CENTRAL_NERVOUS_SYSTEM central_nervous_system "M" glioma astr…
## 2 143B_BONE bone "F" osteosarcoma oste…
## 3 22RV1_PROSTATE prostate "M" carcinoma carc…
## 4 2313287_STOMACH stomach "M" carcinoma aden…
## 5 253JBV_URINARY_TRACT urinary_tract "U" carcinoma tran…
## 6 253J_URINARY_TRACT urinary_tract "" carcinoma tran…ccle_absolute: supplementary information of CCLE samples from Table S2 of the paper
## # A tibble: 6 × 5
## `Cell Line` Lineage Purity Ploidy `Genome Doublings`
## <chr> <chr> <dbl> <dbl> <dbl>
## 1 SKNSH_AUTONOMIC_GANGLIA AUTONOMIC 0.99 2.08 0
## 2 KPNRTBM1_AUTONOMIC_GANGLIA AUTONOMIC 1 1.99 0
## 3 MHHNB11_AUTONOMIC_GANGLIA AUTONOMIC 1 2.16 0
## 4 NH6_AUTONOMIC_GANGLIA AUTONOMIC 1 2.02 0
## 5 IMR32_AUTONOMIC_GANGLIA AUTONOMIC 0.99 2.21 0
## 6 KPNYN_AUTONOMIC_GANGLIA AUTONOMIC 1 1.97 03.4 Zenodo
The immune infiltrations are estimated by immunedeconv package, where 7 types of algorithms (CIBERSORT, CIBERSORT-ABS, EPIC, MCPCOUNTER, QUANTISEQ, TIMER, XCELL) are adopted.
The expression scores for 3 type of gene pathway/signature sets (HALLMARK, KEGG, IOBR) is calculated through ssGSEA method.
3.4.1 tumor infiltration estimations
# TCGA samples
dat1 = load_data("tcga_TIL")
# PCAWG samples
dat2 = load_data("pcawg_TIL")
dat1[1:4,1:4]## # A tibble: 4 × 4
## cell_type `B cell_TIMER` `T cell CD4+_TIMER` `T cell CD8+_TIMER`
## <chr> <dbl> <dbl> <dbl>
## 1 TCGA-OR-A5J1-01 0.108 0.117 0.201
## 2 TCGA-OR-A5J2-01 0.114 0.107 0.213
## 3 TCGA-OR-A5J3-01 0.102 0.106 0.203
## 4 TCGA-OR-A5J5-01 0.102 0.111 0.196- In addition, we also collected expression scores of 160 immune gene signatures across TCGA samples from here.
## Source SetName TCGA-02-0047-01A-01R-1849-01
## Angiogenesis Yasin Angiogenesis 0.1925055
## APM1 Yasin APM1 0.4492857
## APM2 Yasin APM2 0.2437349
## ICS5_score Wolf ICS5_score -1.5192000
## TCGA-02-0055-01A-01R-1849-01
## Angiogenesis 0.09855802
## APM1 0.46742753
## APM2 0.29939846
## ICS5_score 0.617800003.4.2 ssGSEA pathway activities
# TCGA samples
dat1 = load_data("tcga_PW")
# PCAWG samples
dat2 = load_data("pcawg_PW")
dat1[1:4,1:4]## HALLMARK_ADIPOGENESIS HALLMARK_ALLOGRAFT_REJECTION
## TCGA-19-1787-01 0.2857295 0.14104371
## TCGA-S9-A7J2-01 0.2591440 0.03655175
## TCGA-G3-A3CH-11 0.3303460 0.18602627
## TCGA-B5-A5OE-01 0.2692051 0.11716217
## HALLMARK_ANDROGEN_RESPONSE HALLMARK_ANGIOGENESIS
## TCGA-19-1787-01 0.2178818 0.16112190
## TCGA-S9-A7J2-01 0.1826316 0.08244577
## TCGA-G3-A3CH-11 0.2563853 0.15919124
## TCGA-B5-A5OE-01 0.1832165 0.151489923.4.3 other TCGA annotations
- “tcga_stemness”: tumor stemness of TCGA samples from Pan-Cancer Atlas Hub
## sample RNAss EREG.EXPss DNAss EREG-METHss DMPss ENHss
## 1 TCGA-02-0047-01 0.2398426 0.5585645 NA NA NA NA
## 2 TCGA-02-0055-01 0.1878304 0.5743873 NA NA NA NA
## 3 TCGA-02-2483-01 0.4087490 0.7067001 NA NA NA NA
## 4 TCGA-02-2485-01 0.3491451 0.5659132 NA NA NA NA
## 5 TCGA-02-2486-01 0.2498411 0.4618031 NA NA NA NA
## 6 TCGA-04-1348-01 0.5741474 0.4998114 NA NA NA NA- “tcga_tmb”: tumor mutation burden of TCGA samples from Table S1 of the paper
## Cohort Patient_ID Tumor_Sample_ID Silent_per_Mb Non_silent_per_Mb
## 1 ACC TCGA-OR-A5JR TCGA-OR-A5JR-01 0.05168695 0.05168695
## 2 ACC TCGA-OR-A5JH TCGA-OR-A5JH-01 0.10244018 0.15366028
## 3 ACC TCGA-OR-A5JQ TCGA-OR-A5JQ-01 0.08117102 0.16234204
## 4 ACC TCGA-OR-A5L9 TCGA-OR-A5L9-01 0.05354531 0.16063592
## 5 ACC TCGA-OR-A5LA TCGA-OR-A5LA-01 0.05456403 0.19097410
## 6 ACC TCGA-OR-A5LH TCGA-OR-A5LH-01 0.02618618 0.20948946- “tcga_MSI”: tumor microsatellite instability of TCGA samples from Supplementary Data 1 of the paper
## # A tibble: 6 × 22
## Cancer_type Barcode Total_nb_MSI_events MSI_3utr MSI_5utr MSI_exonic
## <chr> <chr> <dbl> <dbl> <dbl> <dbl>
## 1 READ TCGA-DC-4745 10 3 1 6
## 2 READ TCGA-DC-6155 2 0 1 1
## 3 READ TCGA-DC-6681 1 0 0 1
## 4 READ TCGA-EI-6506 0 0 0 0
## 5 READ TCGA-DC-6683 3 0 3 0
## 6 READ TCGA-EI-6885 4 1 1 2
## # ℹ 16 more variables: MSI_noncoding <dbl>, MSI_intronic <dbl>, MSI_mono <dbl>,
## # MSI_di <dbl>, MSI_tri <dbl>, MSI_tetra <dbl>, MSI_3utr_profiled <dbl>,
## # MSI_5utr_profiled <dbl>, MSI_exonic_profiled <dbl>,
## # MSI_noncoding_profiled <dbl>, MSI_intronic_profiled <dbl>,
## # MSI_mono_profiled <dbl>, MSI_di_profiled <dbl>, MSI_tri_profiled <dbl>,
## # MSI_tetra_profiled <dbl>, MSI_category_nb_from_TCGA_consortium <chr>3.4.4 identifier repository
Compile available identifiers of data for each of TPC databases.
- “pancan_identifier_help”: TCGA samples
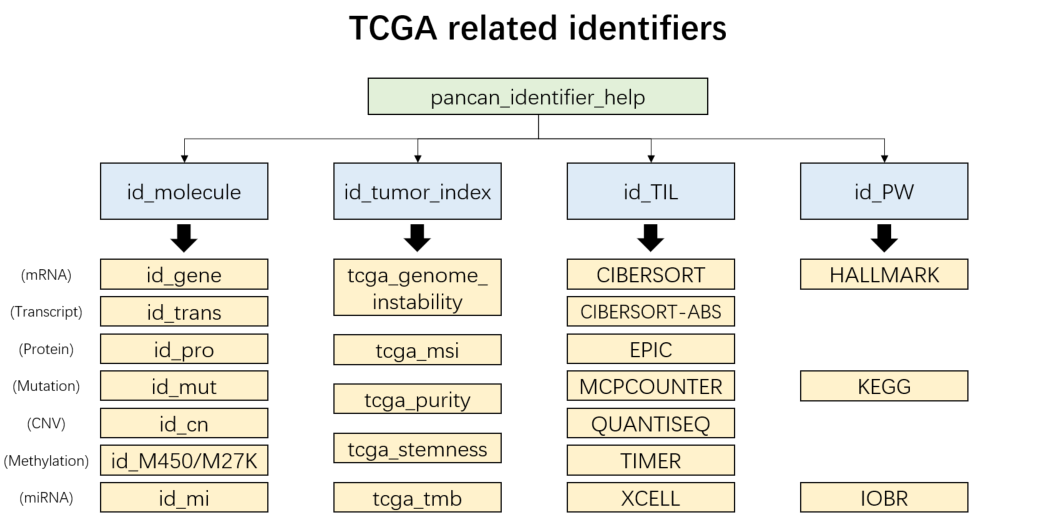
Figure 3.2: TCGA related identifiers
tcga_ids = load_data("pancan_identifier_help")
names(tcga_ids)
# [1] "id_molecule" "id_tumor_index" "id_TIL" "id_PW"
head(tcga_ids$id_molecule$id_gene)
# the key identifier is ususally under "Level3" column## Level2 Level3 Ensembl chrom chromStart chromEnd
## 1 mRNA Expression DDX11L1 ENSG00000223972.5 chr1 11869 14409
## 2 mRNA Expression WASH7P ENSG00000227232.5 chr1 14404 29570
## 3 mRNA Expression MIR6859-1 ENSG00000278267.1 chr1 17369 17436
## 4 mRNA Expression RP11-34P13.3 ENSG00000243485.3 chr1 29554 31109
## 5 mRNA Expression MIR1302-2 ENSG00000274890.1 chr1 30366 30503
## 6 mRNA Expression FAM138A ENSG00000237613.2 chr1 34554 36081
## strand
## 1 +
## 2 -
## 3 -
## 4 +
## 5 +
## 6 -- “pcawg_identifier”: PCAWG samples
- “ccle_identifier”: CCLE samples
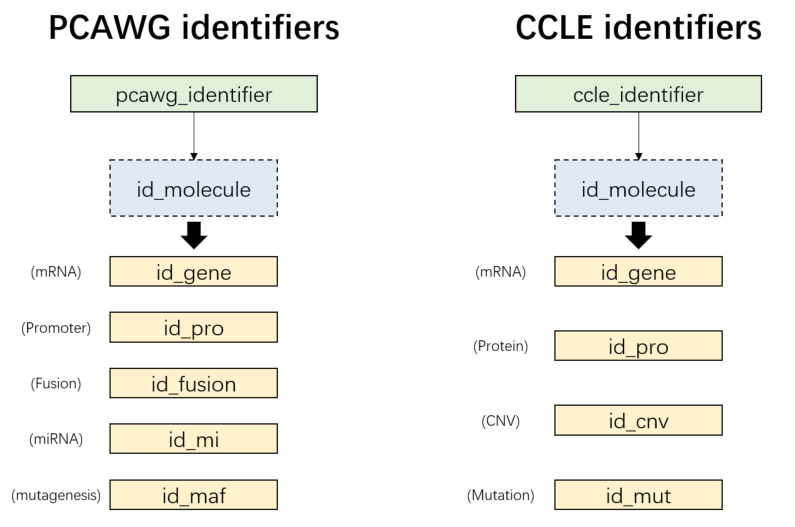
Figure 3.3: PCAWG/CCLE molecular identifiers
pcawg_ids = load_data("pcawg_identifier")
names(pcawg_ids)
# [1] "id_gene" "id_pro" "id_fusion" "id_mi" "id_maf"
head(pcawg_ids$id_pro)
# the key identifier is ususally under "Level3" column## Level2 Level3 gene chrom chromStart chromEnd strand
## 1 Promoter activity prmtr.1 TSPAN6 chrX 99891803 99891803 -
## 2 Promoter activity prmtr.3 TNMD chrX 99839799 99839799 +
## 3 Promoter activity prmtr.6 DPM1 chr20 49575087 49575087 -
## 4 Promoter activity prmtr.7 SCYL3 chr1 169858029 169858029 -
## 5 Promoter activity prmtr.8 SCYL3 chr1 169863093 169863093 -
## 6 Promoter activity prmtr.9 SCYL3 chr1 169863408 169863408 -