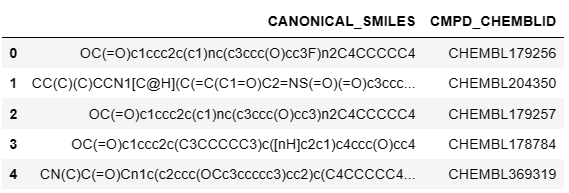
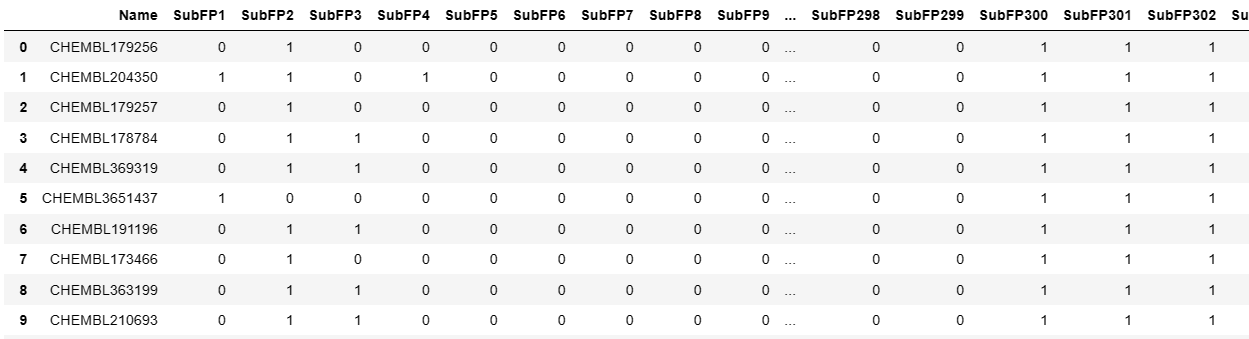
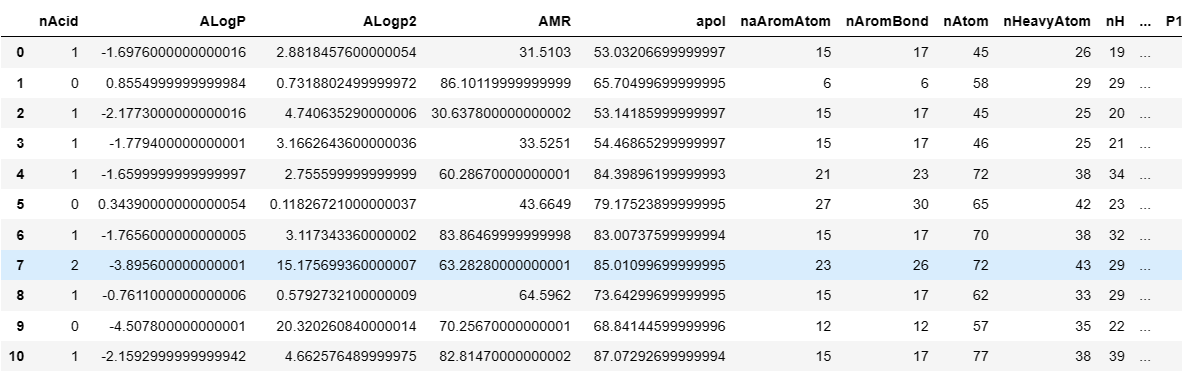
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
|
import glob
xml_files = glob.glob("./fingerprints_xml/*.xml")
xml_files.sort()
xml_files
FP_list = ['AtomPairs2DCount',
'AtomPairs2D',
'EState',
'CDKextended',
'CDK',
'CDKgraphonly',
'KlekotaRothCount',
'KlekotaRoth',
'MACCS',
'PubChem',
'SubstructureCount',
'Substructure']
fp = dict(zip(FP_list, xml_files))
fp
# {'AtomPairs2DCount': './fingerprints_xml/AtomPairs2DFingerprintCount.xml',
# 'AtomPairs2D': './fingerprints_xml/AtomPairs2DFingerprinter.xml',
# 'EState': './fingerprints_xml/EStateFingerprinter.xml',
# 'CDKextended': './fingerprints_xml/ExtendedFingerprinter.xml',
# 'CDK': './fingerprints_xml/Fingerprinter.xml',
# 'CDKgraphonly': './fingerprints_xml/GraphOnlyFingerprinter.xml',
# 'KlekotaRothCount': './fingerprints_xml/KlekotaRothFingerprintCount.xml',
# 'KlekotaRoth': './fingerprints_xml/KlekotaRothFingerprinter.xml',
# 'MACCS': './fingerprints_xml/MACCSFingerprinter.xml',
# 'PubChem': './fingerprints_xml/PubchemFingerprinter.xml',
# 'SubstructureCount': './fingerprints_xml/SubstructureFingerprintCount.xml',
# 'Substructure': './fingerprints_xml/SubstructureFingerprinter.xml'}
|