一、GDSC
GDSC : https://www.cancerrxgene.org/,已上传至阿里云盘
1、原始数据整理
|
|
2、敏感度实验结果
|
|
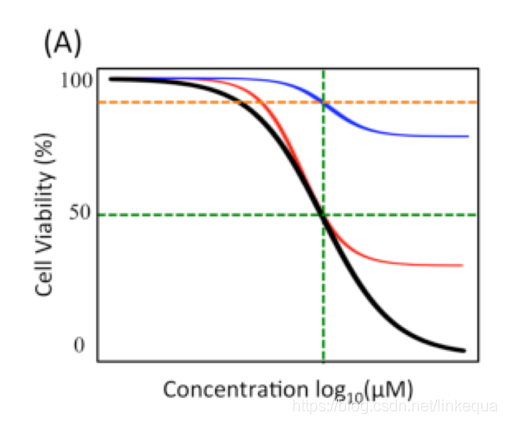
关于IC50与AUC:https://blog.csdn.net/linkequa/article/details/88221975
理论上IC50值或者AUC值越小,表明细胞系对于药物越敏感。
3、药物与细胞系信息
|
|
https://cellmodelpassports.sanger.ac.uk/downloads
在上面网址可下载肿瘤细胞系的多种组学数据,包括转录组、基因组等,有需要时再整理。
二、CTRL
https://ocg.cancer.gov/programs/ctd2/data-portal/ ,已上传至阿里云盘
1、原始数据处理
|
|
v10.D3.area_under_conc_curve.txt:AUCs < 3.5 are considered sensitive to compound treatment, AUCs > 5.5 are considered non-responsive to compound treatment
即AUC值越小,表示细胞系对药物越敏感
- IC50 (half maximal inhibitory concentration)
- EC50 (concentration for 50% of maximal effect,EC50)
2、CTRL v1
|
|
3、CTRL v2
|
|