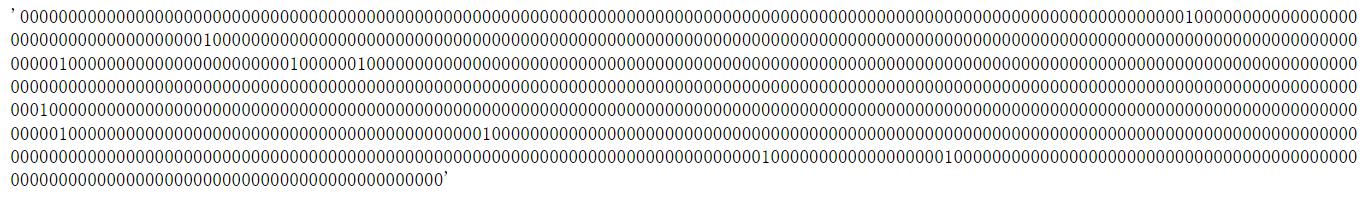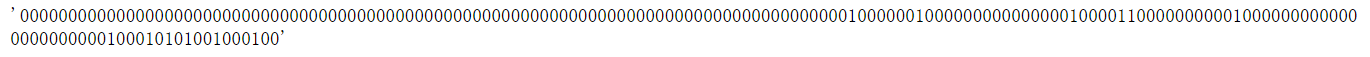1、rdkit#
1
2
3
4
5
6
|
# conda install -c conda-forge rdkit
from rdkit import Chem
from rdkit.Chem import MACCSkeys
from rdkit import DataStructs
from rdkit.Chem import Draw
|
1.1 指纹编码式#
- (1)Topological Fingerprints
1
2
3
4
5
6
7
8
|
m = Chem.MolFromSmiles('CCOC')
# Chem.MolToSmiles(mol)
fp = Chem.RDKFingerprint(m, fpSize=1024)
# fpSize 自定义数目,默认为2048
fp.GetNumBits()
# 1024
fp.ToBitString()
|
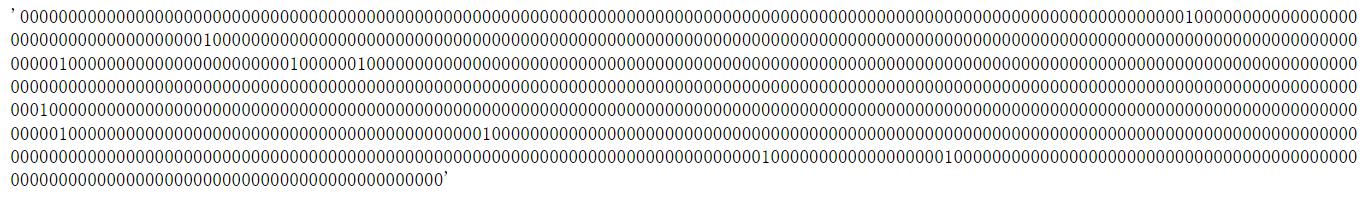
长度为167的分子指纹,每一位都表示一种特定的化学结构特征
https://github.com/openbabel/openbabel/blob/master/data/MACCS.txt
1
2
3
4
5
6
|
from rdkit.Chem import MACCSkeys
fp = MACCSkeys.GenMACCSKeys(m)
fp.GetNumBits()
# 167
fp.ToBitString()
|
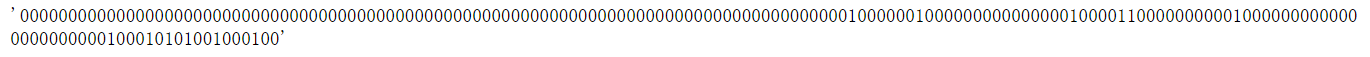
1
2
3
4
5
|
from rdkit.Chem import AllChem
fp = AllChem.GetMorganFingerprintAsBitVect(m1, radius = 6, nBits = 1024)
fp.GetNumBits()
# 1024
fp.ToBitString()
|

1.2 计算相似度#
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
|
m = Chem.MolFromSmiles('CCOC')
fp = MACCSkeys.GenMACCSKeys(m)
m2 = Chem.MolFromSmiles('CCO')
fp2 = MACCSkeys.GenMACCSKeys(m2)
DataStructs.FingerprintSimilarity(fp, fp2)
# 0.5
#默认评价相似度指标为Tanimoto
DataStructs.FingerprintSimilarity(fp, fp2, metric=DataStructs.TanimotoSimilarity)
#还有其它指标
metic_list = ['DataStructs.TanimotoSimilarity',
'DataStructs.DiceSimilarity',
'DataStructs.CosineSimilarity',
'DataStructs.SokalSimilarity',
'DataStructs.RusselSimilarity',
'DataStructs.KulczynskiSimilarity',
'DataStructs.McConnaugheySimilarity']
for i in metic_list:
print(DataStructs.FingerprintSimilarity(fp, fp2, metric=eval(i)))
# 0.5
# 0.6666666666666666
# 0.6735753140545634
# 0.3333333333333333
# 0.041916167664670656
# 0.6805555555555556
# 0.3611111111111111
|
1
2
|
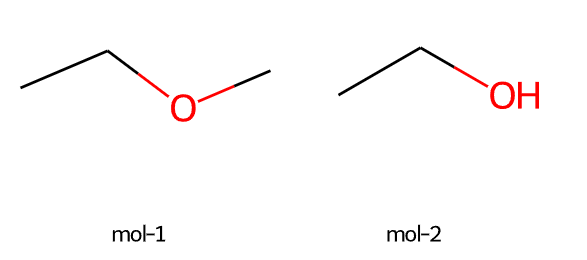
imgs = Draw.MolsToGridImage([m,m2],molsPerRow=2,subImgSize=(200,200), legends=["mol-1","mol-2"])
imgs
|
2、Padel#
1
2
|
pip install padelpy
# pip install pandas
|
示例小分子SMILES文件:https://raw.githubusercontent.com/dataprofessor/data/master/HCV_NS5B_Curated.csv
2.1 指纹编码式#
- (1)12种分子指纹解析文件:https://github.com/dataprofessor/padel/raw/main/fingerprints_xml.zip
1
2
3
4
5
6
7
|
unzip fingerprints_xml.zip
mkdir fingerprints_xml
ls *.xml | while read id; do mv $id ./fingerprints_xml/; done
ls ./fingerprints_xml
# AtomPairs2DFingerprintCount.xml ExtendedFingerprinter.xml KlekotaRothFingerprintCount.xml PubchemFingerprinter.xml
# AtomPairs2DFingerprinter.xml Fingerprinter.xml KlekotaRothFingerprinter.xml SubstructureFingerprintCount.xml
# EStateFingerprinter.xml GraphOnlyFingerprinter.xml MACCSFingerprinter.xml SubstructureFingerprinter.xml
|
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
|
import glob
xml_files = glob.glob("./fingerprints_xml/*.xml")
xml_files.sort()
xml_files
FP_list = ['AtomPairs2DCount',
'AtomPairs2D',
'EState',
'CDKextended',
'CDK',
'CDKgraphonly',
'KlekotaRothCount',
'KlekotaRoth',
'MACCS',
'PubChem',
'SubstructureCount',
'Substructure']
fp = dict(zip(FP_list, xml_files))
fp
|
1
2
3
4
|
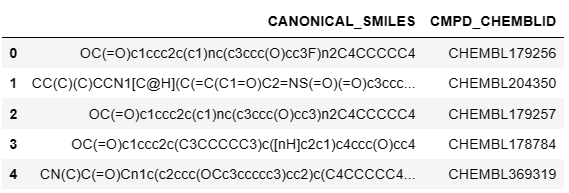
df = pd.read_csv('./HCV_NS5B_Curated.csv')
df[['CANONICAL_SMILES','CMPD_CHEMBLID']].head()
# 保存文件
df2.head(50).to_csv('molecule.smi', sep='\t', index=False, header=False)
|
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
|
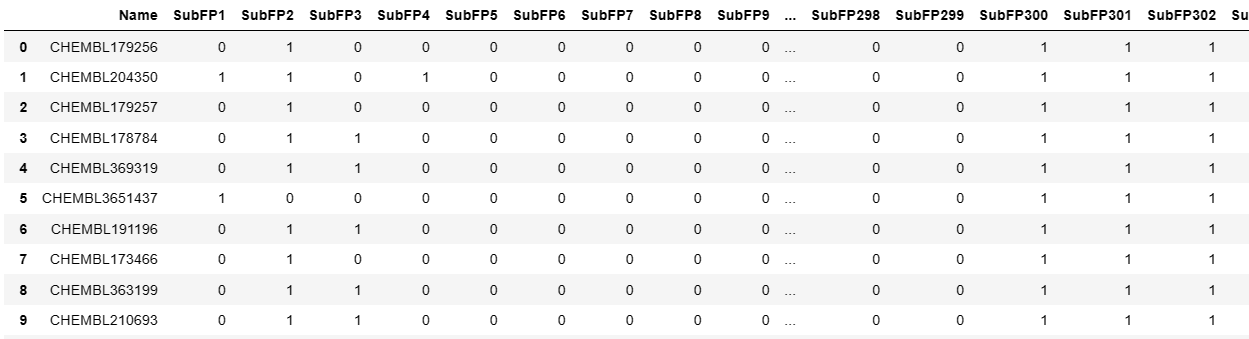
from padelpy import padeldescriptor
# 假设需要转换为Substructure指纹
fingerprint = 'Substructure'
fingerprint_output_file = ''.join([fingerprint,'.csv']) #Substructure.csv 结果文件名
fingerprint_descriptortypes = fp[fingerprint] #解析文件地址
padeldescriptor(mol_dir='molecule.smi',
d_file=fingerprint_output_file, #'Substructure.csv'
#descriptortypes='SubstructureFingerprint.xml',
descriptortypes= fingerprint_descriptortypes,
detectaromaticity=True,
standardizenitro=True,
standardizetautomers=True,
threads=2,
removesalt=True,
log=True,
fingerprints=True)
descriptors = pd.read_csv(fingerprint_output_file)
descriptors
|
2.2 分子描述符#
可根据小分子的SMILES式计算出1875种分子描述符
1
2
3
4
5
6
|
df = pd.read_csv('./HCV_NS5B_Curated.csv')
smi = list(df["CANONICAL_SMILES"][0:50])
descriptors = from_smiles(smi)
descriptors_df = pd.DataFrame(descriptors)
descriptors_df.shape
# (50, 1875)
|
3、mordred#
1
2
3
4
5
6
7
8
9
10
|
# 安装过程,由于版本冲突原因费了一番周折
# (1) python 3.9的环境下
conda install numpy=1.20 networkx=2.3
conda install -c rdkit -c mordred-descriptor mordred
# https://github.com/mordred-descriptor/mordred/issues/84
# (2) 修改DetourMatrix.py模块文件的源代码
# for bcc in networkx.biconnected_component_subgraphs(self.G, False):
# to:
# for bcc in (self.G.subgraph(c) for c in networkx.biconnected_components(self.G)):
|
- 参考github教程,有shell端与python端两种使用方式,如下介绍第一种
1
2
3
|
# 准备好仅包含一列smiles式的文本文件
python -m mordred tmp/canocical_1_col.smi -p 1 --3D \
-o ./mordred_descriptor1826.csv
|
4、ChemDes#
ChemDes网站是由中南大学药学院开发的支持在线计算多种化合物指纹编码式与描述符的网站