1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
|
library(STRINGdb)
library(tidyverse)
string_db <- STRINGdb$new(version="11",
species=9606,
score_threshold=200,
input_directory="")
data(diff_exp_example1)
genes = rbind(head(diff_exp_example1,30),
tail(diff_exp_example1,30))
head(genes)
genes_mapped <- string_db$map(genes, "gene" )
head(genes_mapped)
ppi = string_db$get_interactions(genes_mapped$STRING_id) %>% distinct()
edges = ppi %>%
dplyr::left_join(genes_mapped[,c(1,4)], by=c('from'='STRING_id')) %>%
dplyr::rename(Gene1=gene) %>%
dplyr::left_join(genes_mapped[,c(1,4)], by=c('to'='STRING_id')) %>%
dplyr::rename(Gene2=gene) %>%
dplyr::select(Gene1, Gene2, combined_score)
nodes = genes_mapped %>%
dplyr::filter(gene %in% c(edges$Gene1, edges$Gene2)) %>%
dplyr::mutate(log10P = -log10(pvalue),
direction = ifelse(logFC>0,"Up","Down")) %>%
dplyr::select(gene, log10P, logFC, direction)
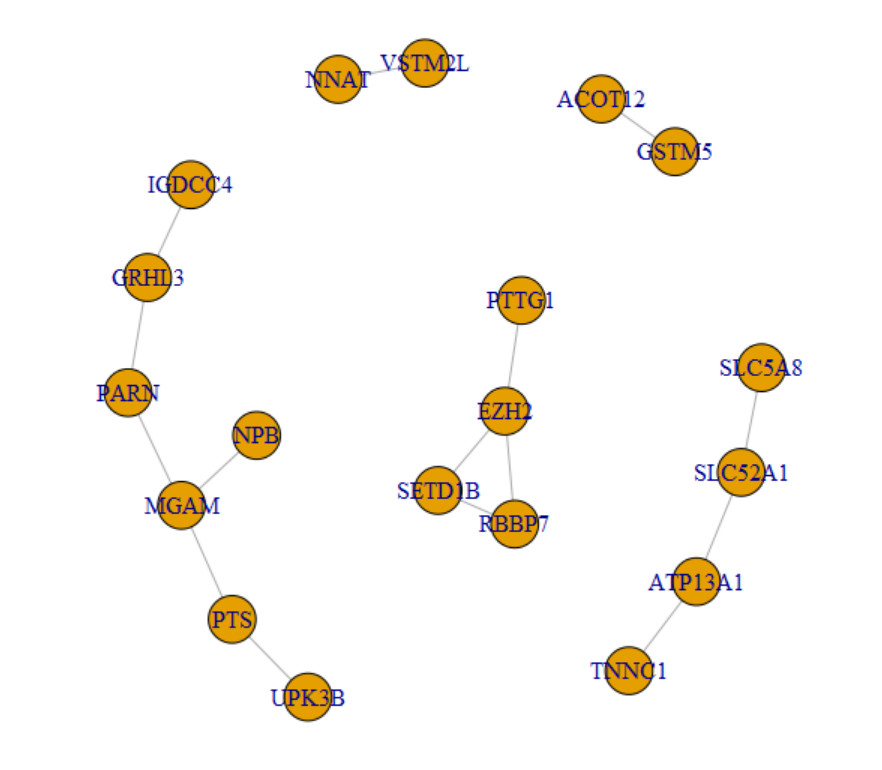
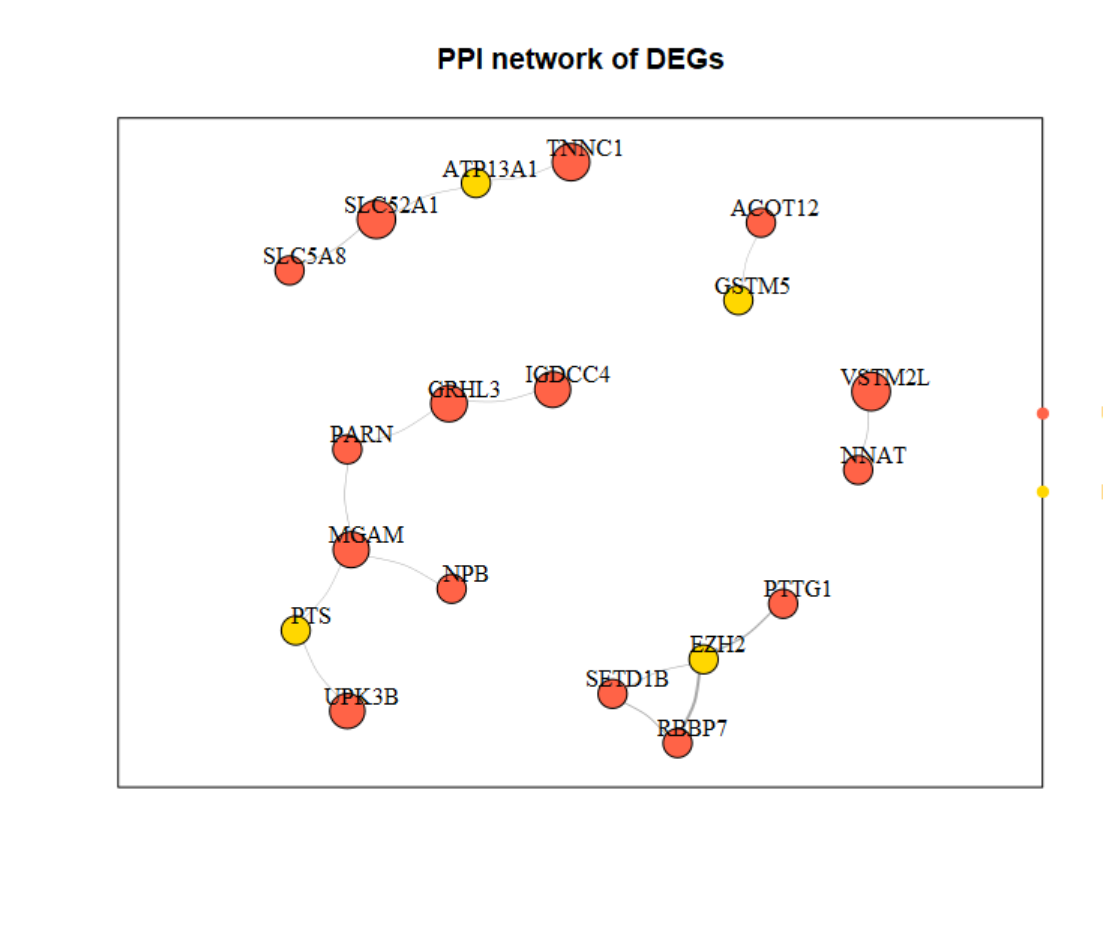
###边信息
head(edges)
# Gene1 Gene2 combined_score
# 1 UPK3B PTS 244
# 2 GSTM5 ACOT12 204
# 3 GRHL3 IGDCC4 238
# 4 TNNC1 ATP13A1 222
# 5 NNAT VSTM2L 281
# 6 EZH2 RBBP7 996
###节点信息
head(nodes)
# gene log10P logFC direction
# 1 VSTM2L 3.992252 3.333461 Up
# 2 TNNC1 3.534468 2.932060 Up
# 3 MGAM 3.515558 2.369738 Up
# 4 IGDCC4 3.290137 2.409806 Up
# 5 UPK3B 3.248490 2.073072 Up
# 6 SLC52A1 3.227019 3.214998 Up
|
](https://s2.loli.net/2022/03/21/hbcfiDXT8gdzUKe.png)
](https://s2.loli.net/2022/03/21/esWG1VLcuyjoQda.png)
](https://s2.loli.net/2022/03/21/TtcP6UIOEVZlaYx.png)
](https://s2.loli.net/2022/03/21/RAaHtrKlFXQYwcD.png)