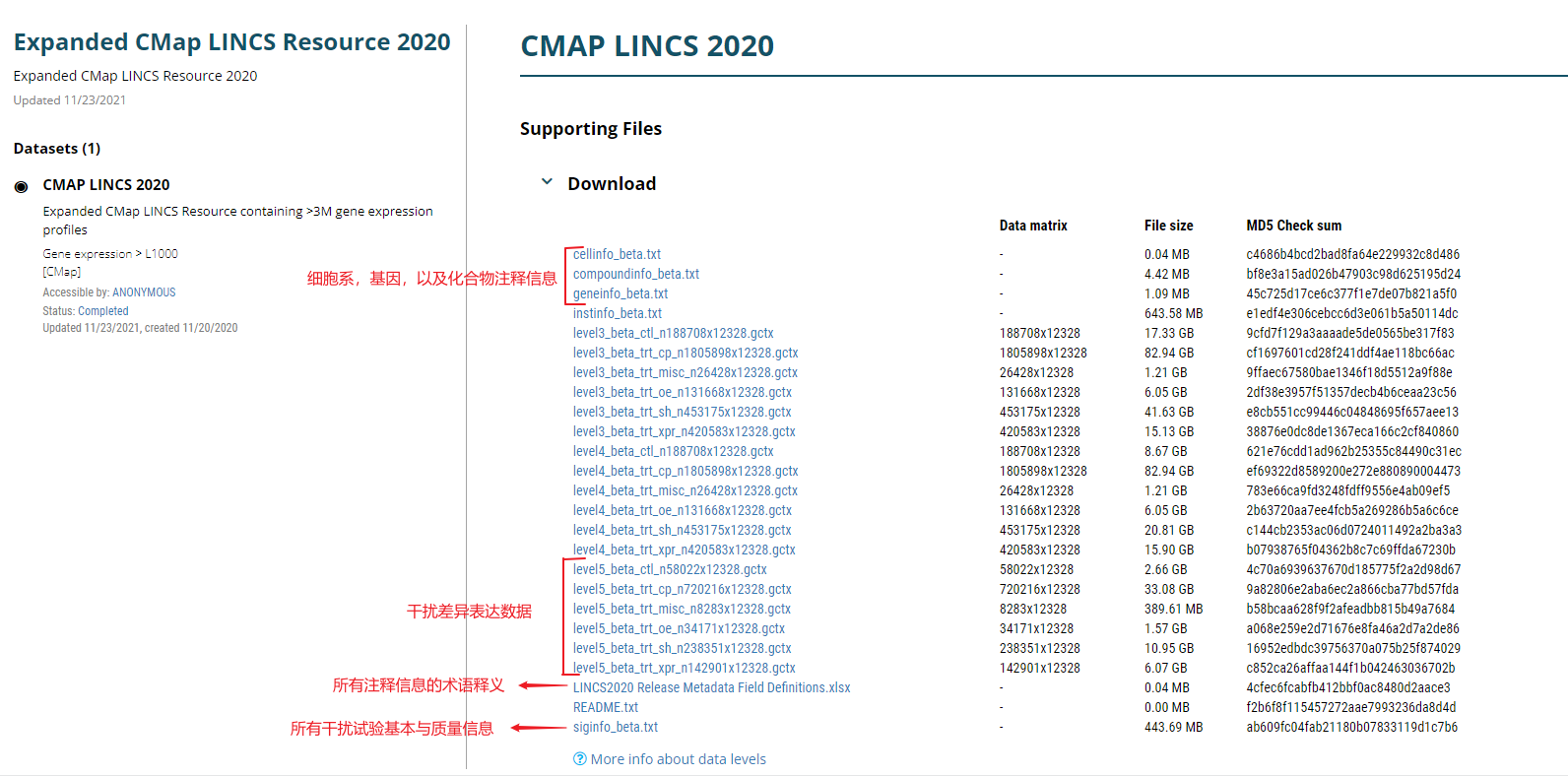
Cmap LINCS计划采用L1000技术进行大规模的细胞系干扰实验测序,得到差异基因。具体可分为Phase-1,Phase-2两个阶段。数据已整理、上传至阿里云盘。本片笔记整理下数据的操作、使用方法。
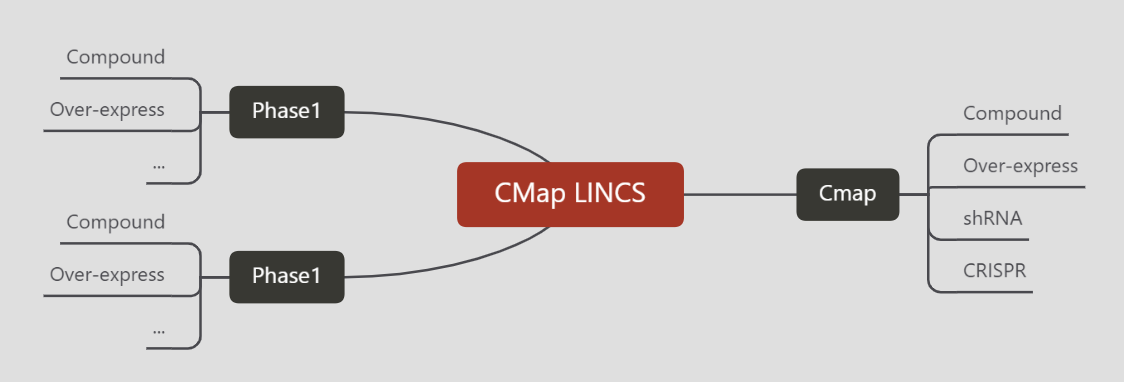
一、CMap#
Compound类型#
- ~4w 化合物作用于240个细胞系的72w+ 次干扰试验
1、差异表达矩阵#
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
|
###根据行列id序号提取指定的差异表达矩阵
###行名表示基因名;列名表示实验id名
gctx_demo = parse_gctx("gctx/level5_beta_trt_cp_n720216x12328.gctx",
cid=1:2, rid=1:2)
gctx_demo@mat
# ABY001_A375_XH:BRD-A61304759:0.625:24 ABY001_A375_XH:BRD-A61304759:0.625:3
# 10 -0.02435 0.2960798
# 100 0.70715 -0.8559232
gctx_demo@rdesc
# id
# 1 10
# 2 100
gctx_demo@cdesc
# id
# 1 ABY001_A375_XH:BRD-A61304759:0.625:24
# 2 ABY001_A375_XH:BRD-A61304759:0.625:3
|
2.1 实验(行)注释–基本+质量信息#
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
|
sig_info = data.table::fread("siginfo_beta.txt",data.table=FALSE)
col_meta <- read_gctx_meta("gctx/level5_beta_trt_cp_n720216x12328.gctx",
dim="col")
table(col_meta[,"id"] %in% sig_info[,"sig_id"])
# TRUE
#720216
sig_sub = subset(sig_info, sig_id %in% col_meta[,"id"])
t(sig_sub[1,])
# bead_batch "b17"
# nearest_dose NA
# pert_dose "100"
# pert_dose_unit "ug/ml"
# pert_idose "100 ug/ml"
# pert_itime "336 h"
# pert_time "336"
# pert_time_unit "h"
# cell_mfc_name "N8"
# pert_mfc_id "BRD-U44432129"
# nsample "4"
# cc_q75 "0.6164"
# ss_ngene "446"
# tas "0.530187"
# pct_self_rank_q25 "0"
# wt "0.26,0.26,0.22,0.26"
# median_recall_rank_spearman "0.925926"
# median_recall_rank_wtcs_50 "1.15741"
# median_recall_score_spearman "0.548655"
# median_recall_score_wtcs_50 "0.705263"
# batch_effect_tstat "-2.31"
# batch_effect_tstat_pct "0.488085"
# is_hiq "1"
# qc_pass "1"
# pert_id "BRD-U44432129"
# sig_id "MET001_N8_XH:BRD-U44432129:100:336"
# pert_type "trt_cp"
# cell_iname "NAMEC8"
# det_wells "H05|H06|H07|H08"
# det_plates "MET001_N8_XH_X1_B17"
# distil_ids "MET001_N8_XH_X1_B17:H05|MET001_N8_XH_X1_B17:H06|MET001_N8_XH_X1_B17:H07|MET001_N8_XH_X1_B17:H08"
# build_name NA
# project_code "MET"
# cmap_name "BRD-U44432129"
# is_exemplar_sig "0"
# is_ncs_sig "0"
# is_null_sig "0"
|
2.2 实验(行)注释–化合物与细胞系#
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
|
cp_meta = data.table::fread("compoundinfo_beta.txt")
t(cp_meta[1,])
# [,1]
# pert_id "BRD-A08715367"
# cmap_name "L-theanine"
# target ""
# moa ""
# canonical_smiles "CCNC(=O)CCC(N)C(O)=O"
# inchi_key "DATAGRPVKZEWHA-UHFFFAOYSA-N"
# compound_aliases "l-theanine"
cell_meta = data.table::fread("cellinfo_beta.txt")
t(cell_meta[1,])
# [,1]
# cell_iname "1HAE"
# cellosaurus_id ""
# donor_age ""
# donor_age_death NA
# donor_disease_age_onset NA
# doubling_time ""
# growth_medium ""
# provider_catalog_id ""
# feature_id ""
# cell_type "normal"
# donor_ethnicity "Unknown"
# donor_sex "Unknown"
# donor_tumor_phase "Unknown"
# cell_lineage "unknown"
# primary_disease "unknown"
# subtype "normal fibroblast sample"
# provider_name ""
# growth_pattern "unknown"
# ccle_name ""
# cell_alias ""
|
2.3 实验(列)注释–基因信息#
1
2
3
4
5
6
7
8
9
10
|
gene_meta = data.table::fread("geneinfo_beta.txt")
t(gene_meta[1,])
# [,1]
# gene_id "750"
# gene_symbol "GAS8-AS1"
# ensembl_id "ENSG00000221819"
# gene_title "GAS8 antisense RNA 1"
# gene_type "ncRNA"
# src "NCBI"
# feature_space "inferred"
|
shRNA类型#
1、差异表达矩阵#
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
|
gctx_demo = parse_gctx("gctx/level5_beta_trt_sh_n238351x12328.gctx",
cid=1:2, rid=1:2)
gctx_demo@mat
# CGS001_A375_96H:A2M:1 CGS001_A375_96H:AARS:1
# 10 -0.3014087 -0.17930678
# 100 0.2917795 0.05233869
gctx_demo@rdesc
# id
# 1 10
# 2 100
gctx_demo@cdesc
# id
# 1 CGS001_A375_96H:A2M:1
# 2 CGS001_A375_96H:AARS:1
|
2.1 实验(行)注释–基本+质量信息#
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
|
sig_info = data.table::fread("siginfo_beta.txt",data.table=FALSE)
col_meta <- read_gctx_meta("gctx/level5_beta_trt_sh_n238351x12328.gctx",
dim="col")
table(col_meta[,"id"] %in% sig_info[,"sig_id"])
# TRUE
# 238351
t(sig_sub[1,])
# 373
# bead_batch "b7"
# nearest_dose NA
# pert_dose NA
# pert_dose_unit ""
# pert_idose ""
# pert_itime "96 h"
# pert_time "96"
# pert_time_unit "h"
# cell_mfc_name "PC3"
# pert_mfc_id "TRCN0000350275"
# nsample "2"
# cc_q75 "0.43"
# ss_ngene "74"
# tas "0.180377"
# pct_self_rank_q25 "0.134953"
# wt "\"\"\"\"\"\"\"\"\"\"\"\"\"\"\"\"\"\"\"\"\"\"\"\"\"\"\"\"\"\"\"\""
# median_recall_rank_spearman "0"
# median_recall_rank_wtcs_50 "0"
# median_recall_score_spearman "0.426979"
# median_recall_score_wtcs_50 "0.697757"
# batch_effect_tstat "11.81"
# batch_effect_tstat_pct "51.0192"
# is_hiq "1"
# qc_pass "1"
# pert_id "TRCN0000350275"
# sig_id "TAK001_PC3_96H:TRCN0000350275:-666"
# pert_type "trt_sh"
# cell_iname "PC3"
# det_wells "P11"
# det_plates "TAK001_PC3_96H_X1_B7_DUO52HI53LO|TAK001_PC3_96H_X2_B7_DUO52HI53LO"
# distil_ids "TAK001_PC3_96H_X1_B7_DUO52HI53LO:P11|TAK001_PC3_96H_X2_B7_DUO52HI53LO:P11"
# build_name NA
# project_code "TAK"
# cmap_name "PIK3CA" #作用对象基因
# is_exemplar_sig "1"
# is_ncs_sig "1"
# is_null_sig "0"
|
(1)其余注释同上
(2)此外还有其它类型干扰(over-expression、CRISPR等);操作基本同上,不再叙述。
二、LINCS#
1、差异表达矩阵#
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
|
###根据行列id序号提取指定的差异表达矩阵
###行名表示基因名;列名表示实验id名
gctx_demo = parse_gctx("phase1/GSE92742_Broad_LINCS_Level5_COMPZ.MODZ_n473647x12328.gctx",
cid=1:2, rid=1:2)
gctx_demo@mat
# CPC005_A375_6H:BRD-A85280935-003-01-7:10
# 5720 0.7737690
# 466 -0.8184680
# CPC005_A375_6H:BRD-A07824748-001-02-6:10
# 5720 -0.6455861
# 466 -0.8107487
###gctx文件里没有更多的注释信息
gctx_demo@rdesc
# id
#1 5720
#2 466
gctx_demo@cdesc
# id
#1 CPC005_A375_6H:BRD-A85280935-003-01-7:10
#2 CPC005_A375_6H:BRD-A07824748-001-02-6:10
|
由于gctx文件太大,可根据下面的注释信息,仅仅提取部分感兴趣的差异表达矩阵。
2.1 实验(行)注释–基本信息#
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
|
sig_info = data.table::fread("phase1/Fine_phase1_sig_info_473647.csv",
data.table = F)
col_meta = read_gctx_meta("phase1/GSE92742_Broad_LINCS_Level5_COMPZ.MODZ_n473647x12328.gctx",
dim="col")
identical(col_meta$id, sig_info$sig_id)
# [1] TRUE
t(sig_info[1,])
# [,1]
# sig_id "CPC005_A375_6H:BRD-A85280935-003-01-7:10" ##实验ID
# pert_id "BRD-A85280935" ##干扰方法ID
# pert_iname "quinpirole" ##干扰物命名(化合物或者基因名)
# pert_type "trt_cp" ##干扰类型(化合物/基因敲除、过表达)
# cell_id "A375" ##细胞系名
# pert_dose "10.0" ##剂量
# pert_dose_unit "µM" ##剂量
# pert_idose "10 µM" ##剂量
# pert_time "6" ##干扰时间
# pert_time_unit "h" ##干扰时间
# pert_itime "6 h" ##干扰时间
# distil_id "CPC005_A375_6H_X1_B3_DUO52HI53LO:K06|CPC005_A375_6H_X2_B3_DUO52HI53LO:K06|CPC005_A375_6H_X3_B3_DUO52HI53LO:K06"
table(sig_info$pert_type)
# ctl_untrt ctl_untrt.cns ctl_vector ctl_vector.cns ctl_vehicle
# 588 30 6826 137 14423
#ctl_vehicle.cns trt_cp trt_lig trt_oe trt_oe.mut
# 61 205034 8256 22205 6
# trt_sh trt_sh.cgs trt_sh.css
# 154993 36720 24368
|
2.2 实验(行)注释–实验质量信息#
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
|
sig_metrics = data.table::fread("phase1/phase1_sig_metrcs.csv",
data.table = F)
col_meta = read_gctx_meta("phase1/GSE92742_Broad_LINCS_Level5_COMPZ.MODZ_n473647x12328.gctx",
dim="col")
sig_metrics = sig_metrics[match(col_meta$id, sig_metrics$sig_id),]
identical(col_meta$id, sig_metrics$sig_id)
# [1] TRUE
t(sig_metrics[1,])
# 53349
# sig_id "CPC005_A375_6H:BRD-A85280935-003-01-7:10"
# pert_id "BRD-A85280935"
# pert_iname "quinpirole"
# pert_type "trt_cp"
# distil_cc_q75 "0.11" # 重复相同实验的结果相似度指标
# distil_ss "2.84895" # 干扰影响强度指标
# ngenes_modulated_up_lm "18"
# ngenes_modulated_dn_lm "15"
# tas "0.101169" # 综合cc_q75与ss的评价指标结果
# pct_self_rank_q25 "7.6087"
# is_exemplar "0"
# distil_nsample "3"
|
2.3 实验(行)注释–干扰物与细胞系#
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
|
###通过pert_id与前面的信息相关联
#(1)干扰物注释:主要针对化合物类干扰实验,提供化合物的CID等信息
pert_info = data.table::fread("phase1/phase1_pert_info.csv")
t(pert_info[1,])
# [,1]
# V1 "1"
# pert_id "56582"
# pert_iname "AKT2"
# pert_type "trt_oe"
# is_touchstone "0"
# inchi_key_prefix "-666"
# inchi_key "-666"
# canonical_smiles "-666"
# pubchem_cid "-666"
#(2)细胞系注释:例如是否为肿瘤细胞系,源自那个部位等
cell_info = data.table::fread("phase1/phase1_cell_info.csv")
t(cell_info[1,])
# [,1]
# cell_id "A375"
# cell_type "cell line"
# base_cell_id "A375"
# precursor_cell_id "-666"
# modification "-666"
# sample_type "tumor"
# primary_site "skin"
# subtype "malignant melanoma"
# original_growth_pattern "adherent"
# provider_catalog_id "CRL-1619"
# original_source_vendor "ATCC"
# donor_age "54"
# donor_sex "F"
# donor_ethnicity "-666"
|
2.4 实验(列)注释–基因信息#
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
|
gene_info = data.table::fread("phase1/Fine_phase1_gene_info_12328.csv",
data.table = F)
row_meta <- read_gctx_meta("phase1/GSE92742_Broad_LINCS_Level5_COMPZ.MODZ_n473647x12328.gctx", dim="row")
identical(as.integer(row_meta$id), gene_info$pr_gene_id)
# [1] TRUE
dim(gene_info)
# [1] 12328 5
t(gene_info[1,])
# 1
# pr_gene_id "5720"
# pr_gene_symbol "PSME1"
# pr_gene_title "proteasome activator subunit 1"
# pr_is_lm "1" # landmark 978
# pr_is_bing "1" # landmark + best inferred gene 10174
|
1、差异表达矩阵#
1
2
3
4
5
6
7
8
|
###根据行列id序号提取指定的差异表达矩阵
###行名表示基因名;列名表示实验id名
gctx_demo = parse_gctx("phase2/GSE70138_Broad_LINCS_Level5_COMPZ_n118050x12328_2017-03-06.gctx",
cid=1:2, rid=1:2)
gctx_demo@mat
# REP.A001_A375_24H:A03 REP.A001_A375_24H:A04
#780 4.2641425 -0.3822108
#7849 0.0572492 0.3043132
|
2.1 实验(行)注释–基本信息#
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
|
sig_info = data.table::fread("phase2/Fine_phase2_sig_info_118050.csv",
data.table = F)
col_meta = read_gctx_meta("phase2/GSE70138_Broad_LINCS_Level5_COMPZ_n118050x12328_2017-03-06.gctx",
dim="col")
identical(col_meta$id, sig_info$sig_id)
# [1] TRUE
t(sig_info[1,])
# 1
# sig_id "REP.A001_A375_24H:A03"
# pert_id "DMSO"
# pert_iname "DMSO"
# pert_type "ctl_vehicle"
# cell_id "A375"
# pert_idose "-666"
# pert_itime "24 h"
# distil_id "REP.A001_A375_24H_X1_B22:A03|REP.A001_A375_24H_X2_B22:A03|REP.A001_A375_24H_X3_B22:A03"
table(sig_info$pert_type)
# ctl_untrt ctl_vector ctl_vehicle trt_cp trt_xpr
# 88 209 6467 107404 3882
|
2.2 实验(行)注释–实验质量信息#
1
2
3
4
5
6
7
8
|
sig_metrics = data.table::fread("phase2/phase2_sig_metrics.csv",
data.table = F)
col_meta = read_gctx_meta("phase1/GSE92742_Broad_LINCS_Level5_COMPZ.MODZ_n473647x12328.gctx",
dim="col")
sig_metrics = sig_metrics[match(col_meta$id, sig_metrics$sig_id),]
identical(col_meta$id, sig_metrics$sig_id)
# [1] TRUE
t(sig_metrics[1,])
|
2.3 实验(行)注释–干扰物与细胞系#
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
|
###通过pert_id与前面的信息相关联
#(1)干扰物注释:主要针对化合物类干扰实验,提供化合物的CID等信息
pert_info = data.table::fread("phase2/phase2_pert_info.csv")
t(pert_info[1,])
# [,1]
# pert_id "BRD-K70792160"
# canonical_smiles "CCN(CC)CCCCN1c2ccccc2Oc2ccc(Cl)cc12"
# inchi_key "GYBXAGDWMCJZJK-UHFFFAOYSA-N"
# pert_iname "10-DEBC"
# pert_type "trt_cp"
# pubchem_id "10521421"
#(2)细胞系注释:例如是否为肿瘤细胞系,源自那个部位等
cell_info = data.table::fread("phase2/phase2_cell_info.csv")
t(cell_info[1,])
# [,1]
# cell_id "A375"
# cell_type "cell line"
# base_cell_id "A375"
# precursor_cell_id "-666"
# modification "-666"
# sample_type "tumor"
# primary_site "skin"
# subtype "malignant melanoma"
# original_growth_pattern "adherent"
# provider_catalog_id "CRL-1619"
# original_source_vendor "ATCC"
# donor_age "54"
# donor_sex "F"
# donor_ethnicity "-666"
|
2.4 实验(列)注释–基因信息#
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
|
gene_info = data.table::fread("phase2/Fine_phase1_gene_info_12328.csv",
data.table = F)
row_meta <- read_gctx_meta("phase2/GSE92742_Broad_LINCS_Level5_COMPZ.MODZ_n473647x12328.gctx", dim="row")
identical(as.integer(row_meta$id), gene_info$pr_gene_id)
# [1] TRUE
dim(gene_info)
# [1] 12328 5
t(gene_info[1,])
# 1
# pr_gene_id "5720"
# pr_gene_symbol "PSME1"
# pr_gene_title "proteasome activator subunit 1"
# pr_is_lm "1" # landmark 978
# pr_is_bing "1" # landmark + best inferred gene 10174
|