1、决策树基础#
1.1 决策树的构成#
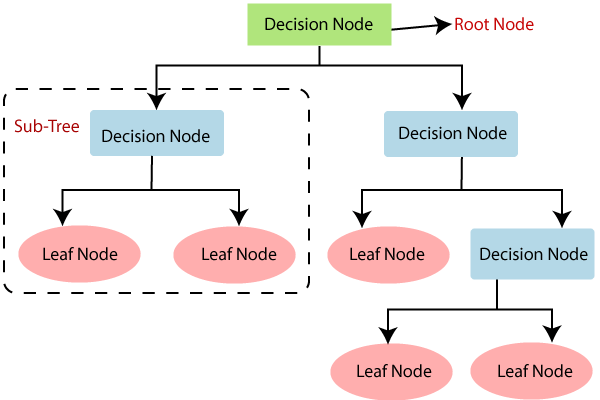
(1)决策树由节点组成,可分为决策节点(Decision tree)与叶节点(leaf node)。
(2)从上到下的第一个节点也称为根节点(Root Node)。根节点到叶节点的最长距离称为树的深度。
(3)对于每一个决策节点,选择样本集的一个最佳预测变量,确定最佳的阈值,进行二分类;最后根据叶节点类别的众数进行分类(均数进行回归)。
1.2 基尼系数#
(1)决策树为了使表示数据杂乱程度的不纯度减小,选择预测变量的最佳cut-off进行分割。当分割出来的每个组中的标签大部分属于同种标签时,不纯度会变小。
- 每一组的复杂度可通过基尼指数表示。其值越小,表示数据纯度越高
- 分割前后的数据复杂度变化可通过基尼增益表示。其值越大,表示数据的纯度变得越高。
(2)基尼指数与基尼增益的计算方法如下图所示:
如公式所示,对于二分类任务的基尼指数取值范围应为0~0.5之间。当等于0时,表示由同一种标签样本组成;当等于0.5时,表示两个标签样本各占一半。

(3)比较不同预测变量的基尼增益,其实就是比较把不同预测变量的最佳分割cut-off下的基尼增益。
具体对于不同类型的预测变量,如上图所示,有不同的选择方法(从若干候选值中选择一个最佳的,作为该预测变量的代表)
1.3 防止决策树过拟合#
(1)直接限制最长深度;
(2)每一次划分所引起的最小性能改进(cp值,类似于基尼增益);
(3)叶节点size:如果划分一个节点会导致叶节点中包含的样本数少于规定值,则不会划分;
(4)决策节点size:如果决策节点少于规定值,则不会被划分。(就被视为了叶节点)
2、决策树mlr建模#
2.1 动物特征数据集#
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
|
data(Zoo, package = "mlbench")
#将逻辑变量(True/False)转为数值型因子变量
Zoo2 = Zoo %>%
dplyr::mutate(across(where(is.logical), as.factor))
str(Zoo2)
# 'data.frame': 101 obs. of 17 variables:
# $ hair : Factor w/ 2 levels "FALSE","TRUE": 2 2 1 2 2 2 2 1 1 2 ...
# $ feathers: Factor w/ 2 levels "FALSE","TRUE": 1 1 1 1 1 1 1 1 1 1 ...
# $ eggs : Factor w/ 2 levels "FALSE","TRUE": 1 1 2 1 1 1 1 2 2 1 ...
# $ milk : Factor w/ 2 levels "FALSE","TRUE": 2 2 1 2 2 2 2 1 1 2 ...
# $ airborne: Factor w/ 2 levels "FALSE","TRUE": 1 1 1 1 1 1 1 1 1 1 ...
# $ aquatic : Factor w/ 2 levels "FALSE","TRUE": 1 1 2 1 1 1 1 2 2 1 ...
# $ predator: Factor w/ 2 levels "FALSE","TRUE": 2 1 2 2 2 1 1 1 2 1 ...
# $ toothed : Factor w/ 2 levels "FALSE","TRUE": 2 2 2 2 2 2 2 2 2 2 ...
# $ backbone: Factor w/ 2 levels "FALSE","TRUE": 2 2 2 2 2 2 2 2 2 2 ...
# $ breathes: Factor w/ 2 levels "FALSE","TRUE": 2 2 1 2 2 2 2 1 1 2 ...
# $ venomous: Factor w/ 2 levels "FALSE","TRUE": 1 1 1 1 1 1 1 1 1 1 ...
# $ fins : Factor w/ 2 levels "FALSE","TRUE": 1 1 2 1 1 1 1 2 2 1 ...
# $ legs : int 4 4 0 4 4 4 4 0 0 4 ...
# $ tail : Factor w/ 2 levels "FALSE","TRUE": 1 2 2 1 2 2 2 2 2 1 ...
# $ domestic: Factor w/ 2 levels "FALSE","TRUE": 1 1 1 1 1 1 2 2 1 2 ...
# $ catsize : Factor w/ 2 levels "FALSE","TRUE": 2 2 1 2 2 2 2 1 1 1 ...
# $ type : Factor w/ 7 levels "mammal","bird",..: 1 1 4 1 1 1 1 4 4 1 ...
##第1~16列为动物特征
##第17列type列为动物名标签
|
2.2 确定预测目标与训练方法#
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
|
#(1)根据1-16列动物特征进行动物类型预测
task_classif = as_task_classif(Zoo2, target = "type")
task_classif$col_roles$stratum = "type"
#使用决策树rpart分类算法
learner = lrn("classif.rpart", predict_type="prob")
learner$param_set
##(2)确定该算法的超参数空间以及遍历方法
search_space = ps(
minsplit = p_int(lower=1,upper=20),
minbucket = p_int(lower=1,upper=20),
cp = p_dbl(lower=0, upper=1),
maxdepth = p_int(lower=1,upper=20)
)
design = expand.grid(minsplit=c(1, 5, 10),
minbucket=c(1, 10, 20),
cp=c(0.1, 0.3, 0.5, 0.9),
maxdepth = c(5, 10, 15)) %>% as.data.table()
##5折交叉验证
resampling = rsmp("cv")
resampling$param_set$values$folds=5
##AUC模型评价指标
measure = msr("classif.acc")
|
2.3 模型训练#
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
|
##创建实例
instance = TuningInstanceSingleCrit$new(
task = task_classif,
learner = learner,
resampling = resampling,
measure = measure,
terminator = trm("none"),
search_space = search_space
)
tuner = tnr("design_points", design = design)
future::plan("multisession")
##超参数优化
tuner$optimize(instance)
as.data.table(instance$archive)[,1:5] %>% head
##(1)确定最佳超参数组合
library(parallel)
library(parallelMap)
#调用多线程
parallelStartSocket(cpus = detectCores())
tunedTreePars <- tuneParams(tree, task = zooTask, # ~30 sec
resampling = cvForTuning,
par.set = treeParamSpace,
control = randSearch)
#停用多线程
parallelStop()
tunedTreePars
# Tune result:
# Op. pars: minsplit=6; minbucket=3; cp=0.0345; maxdepth=9
# mmce.test.mean=0.1200000
##(2)使用上述组合训练模型
tunedTree <- setHyperPars(tree, par.vals = tunedTreePars$x)
tunedTreeModel <- train(tunedTree, zooTask)
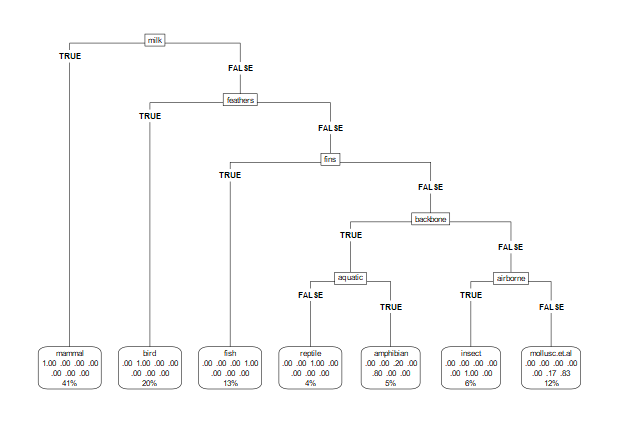
##(3)可视化决策树
treeModelData <- getLearnerModel(tunedTreeModel)
library(rpart.plot)
rpart.plot(treeModelData, roundint = FALSE, type = 5)
|
2.4 嵌套交叉验证#
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
|
#inner
inner <- makeResampleDesc("CV", iters = 5)
#outer
outer <- makeResampleDesc("CV", iters = 3)
treeWrapper <- makeTuneWrapper("classif.rpart", resampling = inner,
par.set = treeParamSpace,
control = randSearch)
parallelStartSocket(cpus = 8)
cvWithTuning <- resample(treeWrapper, zooTask, resampling = outer)
parallelStop()
cvWithTuning
# Resample Result
# Task: zooTib
# Learner: classif.rpart.tuned
# Aggr perf: mmce.test.mean=0.1295306
# Runtime: 13.4196
|
3、bagging与随机森林#
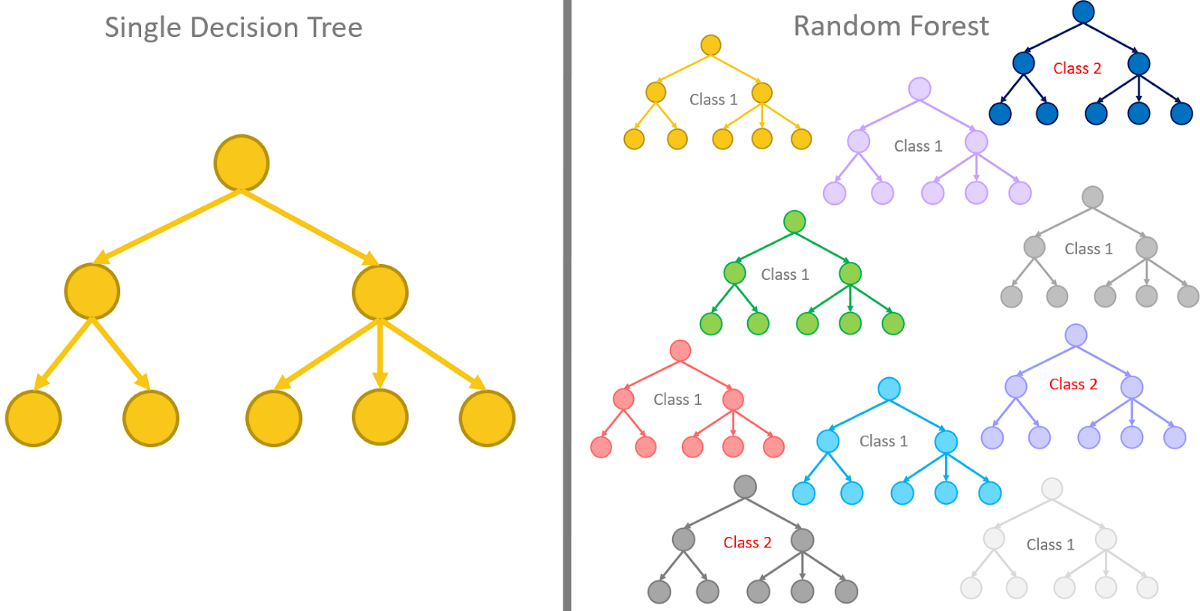
集成学习(Ensemble Learning)是指通过训练多个模型,从而在预测时考虑所有模型的预测结果,可减少单模型的方差。可分为bagging, boosting, stacking三种。虽然集成可用于多种机器学习算法,但还是最常应用于基于树的学习器,例如接下来学到的随机森林就是bagging应用于决策树的经典算法。
由于训练各个子模型的过程理论上互不干扰,因此当训练非常多的子模型时可应用并行化多线程处理。
-
随机森林(Random forest)是Bagging应用于决策树的实现方式。
(1)特征变量抽样:如下图所示,随机森林基于Bagging步骤又多了一步。在上述的第二步结束后,在进行第三步训练每一个决策树时,对特定树的每一个节点都随机选择一定比例的预测变量;然后再从中根据基尼增益选择用于划分的预测变量。
(2)包外(OOB, Out of bag)数据:因为Bagging的自助法抽样,所以每个决策树总会有一些重复样本,而有一定比例的(大部分说法是1/3)样本没有被抽到。换一个角度思考,对于一个样本来说,在随机森林的k个决策树模型中,会有~1/3的决策树训练时没有用到该样本。因此可用来评价、验证模型。
(3)超参数: ①多少个决策树(不考虑计算开销,越多越好);②在每个节点随机采样的特征数量,即上面第一点的特征变量抽样;③叶节点的最小样本数等与决策树类似的超参数。
(4)降低方差:bagging适合用于决策树算法,因为bagging的集成方法有利于降低不稳定的强学习器base learnner,比如决策树算法一般预测方差很大。
4、随机森林mlr建模#
仍使用上面训练决策树所用到的模型
4.1 动物特征训练数据集#
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
|
library(mlr3verse)
library(tidyverse)
data(Zoo, package = "mlbench")
Zoo2 = Zoo %>%
dplyr::mutate(across(where(is.logical), as.factor))
str(Zoo2)
# 'data.frame': 101 obs. of 17 variables:
# $ hair : Factor w/ 2 levels "FALSE","TRUE": 2 2 1 2 2 2 2 1 1 2 ...
# $ feathers: Factor w/ 2 levels "FALSE","TRUE": 1 1 1 1 1 1 1 1 1 1 ...
# $ eggs : Factor w/ 2 levels "FALSE","TRUE": 1 1 2 1 1 1 1 2 2 1 ...
# $ milk : Factor w/ 2 levels "FALSE","TRUE": 2 2 1 2 2 2 2 1 1 2 ...
# $ airborne: Factor w/ 2 levels "FALSE","TRUE": 1 1 1 1 1 1 1 1 1 1 ...
# $ aquatic : Factor w/ 2 levels "FALSE","TRUE": 1 1 2 1 1 1 1 2 2 1 ...
# $ predator: Factor w/ 2 levels "FALSE","TRUE": 2 1 2 2 2 1 1 1 2 1 ...
# $ toothed : Factor w/ 2 levels "FALSE","TRUE": 2 2 2 2 2 2 2 2 2 2 ...
# $ backbone: Factor w/ 2 levels "FALSE","TRUE": 2 2 2 2 2 2 2 2 2 2 ...
# $ breathes: Factor w/ 2 levels "FALSE","TRUE": 2 2 1 2 2 2 2 1 1 2 ...
# $ venomous: Factor w/ 2 levels "FALSE","TRUE": 1 1 1 1 1 1 1 1 1 1 ...
# $ fins : Factor w/ 2 levels "FALSE","TRUE": 1 1 2 1 1 1 1 2 2 1 ...
# $ legs : int 4 4 0 4 4 4 4 0 0 4 ...
# $ tail : Factor w/ 2 levels "FALSE","TRUE": 1 2 2 1 2 2 2 2 2 1 ...
# $ domestic: Factor w/ 2 levels "FALSE","TRUE": 1 1 1 1 1 1 2 2 1 2 ...
# $ catsize : Factor w/ 2 levels "FALSE","TRUE": 2 2 1 2 2 2 2 1 1 1 ...
# $ type : Factor w/ 7 levels "mammal","bird",..: 1 1 4 1 1 1 1 4 4 1 ...
##第1~16列为动物特征
##第17列为动物名标签
|
4.2 确定预测目标与训练方法#
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
|
#(1)确定预测目标,根据1~16列数据预测动物类别
task_classif = as_task_classif(Zoo2, target = "type")
task_classif$col_roles$stratum = "type"
#(2)指定随机森林分类算法
learner = lrn("classif.rpart", predict_type="prob")
learner$param_set
#(3)超参数组合以及遍历方法
search_space = ps(
num.trees = p_int(lower=1,upper=1000),
mtry = p_int(lower=6,upper=12),
min.node.size = p_dbl(lower=1, upper=5),
max.depth = p_int(lower=1,upper=20)
)
design = expand.grid(num.trees=c(300, 500, 1000),
mtry=c(6, 9, 12),
min.node.size=c(1,2,3,4,5),
max.depth = c(3, 6, 9)) %>% as.data.table()
#(4)交叉验证方法与模型评价指标
resampling = rsmp("cv")
measure = msr("classif.acc")
|
4.3 模型训练优化超参数#
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
|
##创建实例
instance = TuningInstanceSingleCrit$new(
task = task_classif,
learner = learner,
resampling = resampling,
measure = measure,
terminator = trm("none"),
search_space = search_space
)
tuner = tnr("design_points", design = design)
##优化超参数
tuner$optimize(instance)
as.data.table(instance$archive)[,1:5]
# num.trees mtry min.node.size max.depth classif.acc
# 1: 300 6 1 3 0.9311764
# 2: 500 6 1 3 0.9200653
# 3: 1000 6 1 3 0.9200653
# 4: 300 9 1 3 0.8814005
# 5: 500 9 1 3 0.9004481
# ---
# 131: 500 9 5 9 0.9718615
# 132: 1000 9 5 9 0.9718615
# 133: 300 12 5 9 0.9623377
# 134: 500 12 5 9 0.9623377
# 135: 1000 12 5 9 0.9718615
instance$result_learner_param_vals
# $num.threads
# [1] 1
# $num.trees
# [1] 300
# $mtry
# [1] 12
# $min.node.size
# [1] 4
# $max.depth
# [1] 6
instance$result_y
#classif.acc
#0.9813853
|
4.4 使用最佳超参数建模#
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
|
learner$param_set$values = instance$result_learner_param_vals
learner$train(task_classif)
learner$model
# Ranger result
# Call:
# ranger::ranger(dependent.variable.name = task$target_names, data = task$data(), probability = self$predict_type == "prob", case.weights = task$weights$weight, num.threads = 1L, num.trees = 300L, mtry = 12L, min.node.size = 4L, max.depth = 6L)
# Type: Probability estimation
# Number of trees: 300
# Sample size: 101
# Number of independent variables: 16
# Mtry: 12
# Target node size: 4
# Variable importance mode: none
# Splitrule: gini
# OOB prediction error (Brier s.): 0.04122035
predict(learner$model, task_classif)
prediction = learner$predict(task_classif)
prediction$confusion
# truth
# response mammal bird reptile fish amphibian insect mollusc.et.al
# mammal 41 0 0 0 0 0 0
# bird 0 20 0 0 0 0 0
# reptile 0 0 5 0 0 0 0
# fish 0 0 0 13 0 0 0
# amphibian 0 0 0 0 4 0 0
# insect 0 0 0 0 0 8 0
# mollusc.et.al 0 0 0 0 0 0 10
prediction$score(msrs(c("classif.acc","classif.ce")))
# classif.acc classif.ce
# 1 0
|



