1
2
|
library(mlr3verse)
library(tidyverse)
|
1、Task训练数据与目的#
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
|
## 分类任务
task_classif = as_task_classif(data, target = "col_target")
#根据预测结果又可分为:twoclass二分类, multiclass多分类
## 回归任务
task_regr = as_task_regr(data, target = "col_target")
task$ncol
task$nrow
task$feature_names
task$feature_types
task$target_names
task$task_type
task$data()
task$col_roles
|
2、Learner 机器学习算法#
1
2
3
4
|
mlr_learners$keys()
ALL_mlr_lrn = as.data.table(mlr_learners)
dim(ALL_mlr_lrn)
# [1] 135 7
|
2.1 分类算法#
1
2
3
4
5
6
7
8
9
10
11
12
13
|
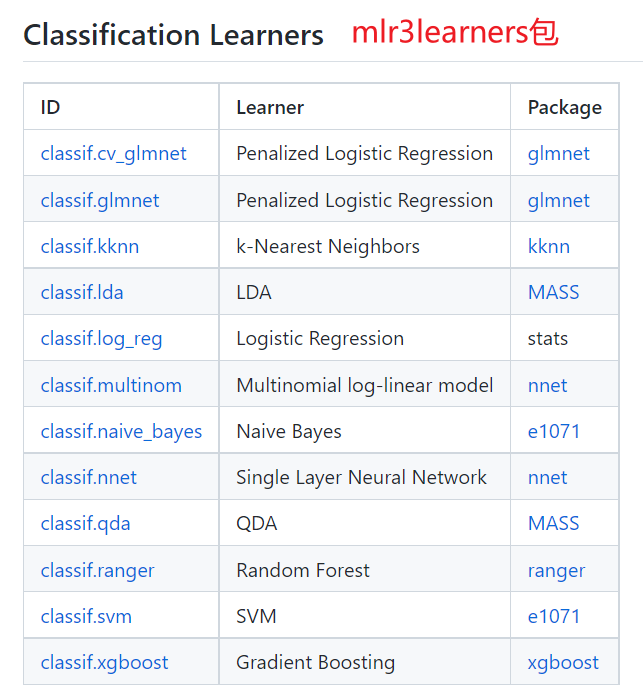
## mlr3extralearners包所支持的全部分类机器学习算法
mlr_learners$keys() %>% grep("classif", . , value=TRUE)
# [1] "classif.AdaBoostM1" "classif.bart" "classif.C50" "classif.catboost"
# [5] "classif.cforest" "classif.ctree" "classif.cv_glmnet" "classif.debug"
# [9] "classif.earth" "classif.featureless" "classif.fnn" "classif.gam"
# [13] "classif.gamboost" "classif.gausspr" "classif.gbm" "classif.glmboost"
# [17] "classif.glmnet" "classif.IBk" "classif.J48" "classif.JRip"
# [21] "classif.kknn" "classif.ksvm" "classif.lda" "classif.liblinear"
# [25] "classif.lightgbm" "classif.LMT" "classif.log_reg" "classif.lssvm"
# [29] "classif.mob" "classif.multinom" "classif.naive_bayes" "classif.nnet"
# [33] "classif.OneR" "classif.PART" "classif.qda" "classif.randomForest"
# [37] "classif.ranger" "classif.rfsrc" "classif.rpart" "classif.svm"
# [41] "classif.xgboost"
|
2.2 回归算法#
1
2
3
4
5
6
7
8
9
10
11
|
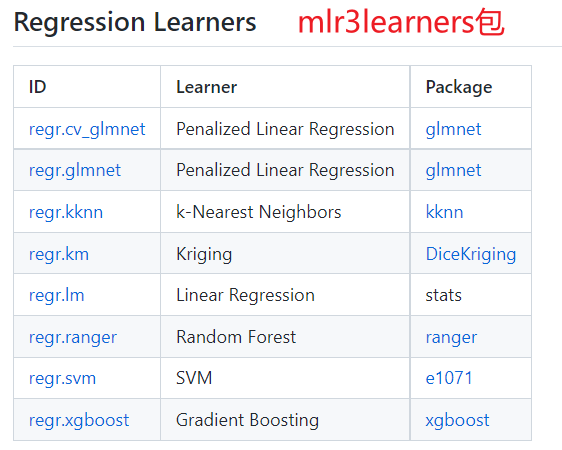
## mlr3extralearners包所支持的全部回归机器学习算法
mlr_learners$keys() %>% grep("regr", . , value=TRUE)
# [1] "regr.bart" "regr.catboost" "regr.cforest" "regr.ctree"
# [5] "regr.cubist" "regr.cv_glmnet" "regr.debug" "regr.earth"
# [9] "regr.featureless" "regr.fnn" "regr.gam" "regr.gamboost"
# [13] "regr.gausspr" "regr.gbm" "regr.glm" "regr.glmboost"
# [17] "regr.glmnet" "regr.IBk" "regr.kknn" "regr.km"
# [21] "regr.ksvm" "regr.liblinear" "regr.lightgbm" "regr.lm"
# [25] "regr.lmer" "regr.M5Rules" "regr.mars" "regr.mob"
# [29] "regr.randomForest" "regr.ranger" "regr.rfsrc" "regr.rpart"
# [33] "regr.rvm" "regr.svm" "regr.xgboost"
|
2.3 算法超参数#
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
|
learner = lrn("classif.kknn")
learner
# <LearnerClassifKKNN:classif.kknn>
# * Model: -
# * Parameters: k=7
# * Packages: mlr3, mlr3learners, kknn
# * Predict Type: response
# * Feature types: logical, integer, numeric, factor, ordered
# * Properties: multiclass, twoclass
## 对于分类算法的预测结果默认的Predict Type为response
## 设置为prob,则会返回属于每一类别的概率
# learner = lrn("classif.kknn", predict_type = "prob")
#算法的超参数
learner$param_set
# <ParamSet>
# id class lower upper nlevels default value
# 1: k ParamInt 1 Inf Inf 7 7
# 2: distance ParamDbl 0 Inf Inf 2
# 3: kernel ParamFct NA NA 10 optimal
# 4: scale ParamLgl NA NA 2 TRUE
# 5: ykernel ParamUty NA NA Inf
# 6: store_model ParamLgl NA NA 2 FALSE
#修改设置超参数
learner$param_set$values
# $k
# [1] 7
learner$param_set$values$k = 3
|
3、交叉验证评价模型#
1
2
3
4
|
ALL_mlr_resample = as.data.table(mlr_resamplings)
mlr_resamplings$keys()
# [1] "bootstrap" "custom" "custom_cv" "cv" "holdout" "insample"
# [7] "loo" "repeated_cv" "subsampling"
|
3.1 评价指标#
mlr3包提供了评价分类、回归模型的多种评价指标。
其中对于分类模型,根据是多分类还是二分类、预测结果形式是response还是prop有不同的评价指标,注意区分。
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
|
mlr_measures$keys()
ALL_mlr_msr = as.data.table(mlr_measures)
head(ALL_mlr_msr)
# key label task_type packages predict_type task_properties
# 1: aic Akaika Information Criterion <NA> mlr3 response
# 2: bic Bayesian Information Criterion <NA> mlr3 response
# 3: classif.acc Classification Accuracy classif mlr3,mlr3measures response
# 4: classif.auc Area Under the ROC Curve classif mlr3,mlr3measures prob twoclass
# 5: classif.bacc Balanced Accuracy classif mlr3,mlr3measures response
# 6: classif.bbrier Binary Brier Score classif mlr3,mlr3measures prob twoclass
ALL_mlr_msr %>%
dplyr::filter(task_type=="classif",
predict_type=="response",
task_properties!="twoclass") %>% .[,c(1,2)]
# key label
# 1: classif.acc Classification Accuracy
# 2: classif.bacc Balanced Accuracy
# 3: classif.ce Classification Error
# 4: classif.costs Cost-sensitive Classification
|
3.2 留出法交叉验证#
将数据分为训练集与测试集。首先在训练集训练模型参数,然后在测试集测试模型性能。
(1)手动拆分数据集#
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
|
task = as_task_classif(diabetes, target = "class")
split = partition(task, ratio = 0.6, stratify = T)
str(split)
# List of 2
# $ train: int [1:88] 2 3 5 6 7 9 10 11 12 13 ...
# $ test : int [1:57] 1 4 8 17 19 20 21 22 23 24 ...
learner = lrn("classif.kknn")
#先用训练集训练模型
learner$train(task, row_ids = split$train)
#再在测试集评价模型
prediction = learner$predict(task, row_ids = split$test)
as.data.table(prediction) %>% head
# row_ids truth response
# 1: 2 Normal Normal
# 2: 5 Normal Normal
# 3: 7 Normal Normal
#指标评价
prediction$score(msr("classif.acc"))
|
(2)使用holdout验证方法(推荐)#
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
|
task$col_roles$stratum = "class"
#对于交叉验证,分训练集与测试集时根据 class列进行分层抽样
resampling = rsmp("holdout")
print(resampling)
resampling$param_set$values
# $ratio
# [1] 0.6666667
# 默认2/3 训练集 1/3测试集
# split = resampling$instantiate(task)
# str(split$instance)
# # List of 2
# # $ train: int [1:97] 1 3 4 5 6 7 12 14 15 17 ...
# # $ test : int [1:48] 2 8 9 10 11 13 16 21 23 28 ...
rr = resample(task, learner, resampling)
str(rr)
# Classes 'ResampleResult', 'R6' <ResampleResult>
rr$prediction()
# <PredictionClassif> for 48 observations:
## 对于测试集的预测结果
rr$prediction() %>% as.data.table() %>% head
# row_ids truth response
# 1: 2 Normal Normal
# 2: 8 Normal Normal
# 3: 9 Normal Normal
rr$score(msr("classif.acc"))[,c(-1,-3,-5,-8)]
# task_id learner_id resampling_id iteration classif.acc
# 1: diabetes classif.kknn holdout 1 0.8541667
rr$score(msrs(c("classif.acc","classif.bacc","classif.ce")))[,c(-1,-3,-5,-8)]
# task_id learner_id resampling_id iteration classif.acc classif.bacc classif.ce
# 1: diabetes classif.kknn holdout 1 0.8541667 0.8397436 0.1458333
|
3.3 K折交叉验证#
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
|
resampling = rsmp("cv")
print(resampling)
resampling$param_set$values
# $folds
# [1] 10
# 默认10折交叉验证
rr = resample(task, learner, resampling)
str(rr)
# Classes 'ResampleResult', 'R6' <ResampleResult>
rr$prediction()
# <PredictionClassif> for 145 observations:
## 对于测试集的预测结果
rr$prediction() %>% as.data.table() %>% head
##性能评价
rr$score(msr("classif.acc"))[,c(-1,-3,-5,-8)]
# task_id learner_id resampling_id iteration classif.acc
# 1: diabetes classif.kknn cv 1 1.0000000
# 2: diabetes classif.kknn cv 2 0.9333333
# 3: diabetes classif.kknn cv 3 0.8000000
# 4: diabetes classif.kknn cv 4 0.8000000
# 5: diabetes classif.kknn cv 5 0.8666667
# 6: diabetes classif.kknn cv 6 0.9285714
# 7: diabetes classif.kknn cv 7 0.9285714
# 8: diabetes classif.kknn cv 8 0.9285714
# 9: diabetes classif.kknn cv 9 0.9285714
# 10: diabetes classif.kknn cv 10 0.9285714
rr$aggregate(msr("classif.acc"))
# classif.acc
# 0.9042857
rr$aggregate(msrs(c("classif.acc","classif.bacc","classif.ce")))
# classif.acc classif.bacc classif.ce
# 0.90428571 0.88711520 0.09571429
|
4、超参数优化#
每种机器学习算法都有特定含义的超参数。
超参数优化是指寻找一个超参数使得模型的性能相对来说由于其它超参数。
首先需要选择候选超参数,其次需要定义模型性能的评价方式与指标。
4.1 简单流程#
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
|
task = as_task_classif(diabetes, target = "class")
learner = lrn("classif.kknn")
##定义候选超参数范围--3、4、5、6、7、8、9、10
search_space <- ps(
k = p_int(lower = 3, upper = 10)
)
##定义模型评价方式--10折交叉验证
resampling = rsmp("cv")
##定义模型评价指标--分类错误率
measure = msr("classif.ce")
##根据可用的预算提前终止训练
terminator = trm("none")
instance = TuningInstanceSingleCrit$new(
task = task,
learner = learner,
resampling = resampling,
measure = measure,
terminator = trm("none"),
search_space = search_space
)
tuner = tnr("grid_search") #默认resolution=10
tuner$optimize(instance)
instance$archive %>% as.data.table() %>% .[,c(1,2)]
# k classif.ce
# 1: 4 0.10333333
# 2: 6 0.09619048
# 3: 9 0.08952381
# 4: 3 0.08952381
# 5: 10 0.10285714
# 6: 5 0.10333333
# 7: 8 0.08952381
# 8: 7 0.09619048
##最优超参数
instance$result_learner_param_vals
# $k
# [1] 8
##最优超参数的模型指标
instance$result_y
# classif.ce
# 0.07619048
|
4.2 设置超参数空间#
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
|
# 整型
p_int(lower=1,upper=10)
# 小数
p_dbl(lower=0.01,upper=1)
# 字符类型
p_fct(c("a", "b", "c"))
# 逻辑值
p_lgl()
search_space <- ps(
hp1 = p_int(lower=1,upper=10),
hp2 = p_dbl(lower=0.01,upper=1),
hp3 = p_fct(c("a", "b", "c"))
)
# <ParamSet>
# id class lower upper nlevels default value
# 1: hp1 ParamInt 1.00 10 10 <NoDefault[3]>
# 2: hp2 ParamDbl 0.01 1 Inf <NoDefault[3]>
# 3: hp3 ParamFct NA NA 3 <NoDefault[3]>
|
如上,当设置小数类型的超参数时,默认有无限种可能。因此需要resolution参数限定。
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
|
# (1)resolution 默认以上下边界值的n个等距值
# 3*3*3 = 27
generate_design_grid(search_space, resolution = 3)
# <Design> with 27 rows:
# hp1 hp2 hp3
# 1: 1 0.010 a
# 2: 1 0.010 b
# 3: 1 0.010 c
# (2)param_resolutions为不同的超参数设置不同的resolution
generate_design_grid(search_space,
param_resolutions = c(hp1 = 5, hp2 = 3))
# <Design> with 45 rows:
# hp1 hp2 hp3
# 1: 1 0.010 a
# 2: 1 0.010 b
# 3: 1 0.010 c
# (3) trafo定义不等间距的超参数范围
search_space = ps(
hp1 = p_int(-1,2, trafo = function(x) 10^x),
hp2 = p_int(1,5, trafo = function(x) 2^x)
)
data.table::rbindlist(generate_design_grid(search_space,
param_resolutions = c(hp1 = 4, hp2 = 5))$transpose())
# hp1 hp2
# 1: 0.1 2
# 2: 0.1 4
# 3: 0.1 8
# 4: 0.1 16
# 5: 0.1 32
|
4.3 设置遍历方式#
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
|
as.data.table(mlr_tuners)[,c(1,2)]
# key label
# 1: cmaes Covariance Matrix Adaptation Evolution Strategy
# 2: design_points Design Points
# 3: gensa Generalized Simulated Annealing
# 4: grid_search Grid Search
# 5: irace Iterated Racing
# 6: nloptr Non-linear Optimization
# 7: random_search Random Search
## grid_search 会将每一种超参数组合计算一遍
## design_points仅遍历自定义的超参数组合范围(推荐)
##(1) 上面提到的resolution方式
search_space = ps(
k = p_int(lower = 3, upper = 50)
)
design = generate_design_grid(search_space, resolution = 5)
design$data
# k
# 1: 3
# 2: 14
# 3: 26
# 4: 38
# 5: 50
tuner = tnr("design_points", design = design$data)
tuner$optimize(instance)
# 需要注意design的候选超参数必须包括在instance的search_space范围内
##(2) 完全自定义的获选超参数
design = data.frame(k=c(3,5,7,9)) %>% as.data.table()
tuner = tnr("design_points", design = design)
tuner$optimize(instance)
|
4.4 关于terminator#
当提供的候选超参数组合过多,占据太多计算资源时,设置TuningInstanceSingleCrit$new()的terminator参数提前种终止遍历。
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
|
as.data.table(mlr_terminators)
# key label properties unit
# 1: clock_time Clock Time single-crit,multi-crit seconds
# 2: combo Combination single-crit,multi-crit percent
# 3: evals Number of Evaluation single-crit,multi-crit evaluations
# 4: none None single-crit,multi-crit percent
# 5: perf_reached Performance Level single-crit percent
# 6: run_time Run Time single-crit,multi-crit seconds
# 7: stagnation Stagnation single-crit percent
# 8: stagnation_batch Stagnation Batch single-crit percent
# clock_time--在给定时间后终止
# evals--在给定的迭代量之后终止
# perf_reached--在达到特定性能后终止
# stagnation--当优化没有改善时终止
# none--不设置终止条件
|
5、训练最终模型与预测新数据#
根据超参数优化结果,基于全部数据,选用最佳的超参数训练得到最终的机器学习模型
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
|
task = as_task_classif(diabetes, target = "class")
learner = lrn("classif.kknn")
learner$param_set$values$k = 8
learner$train(task)
#查看模型
learner$model
#使用模型预测新数据
new_data = data.frame(glucose=c(200,300),
insulin=c(500,1000),
sspg=c(100,50))
new_data
# glucose insulin sspg
# 1 200 500 100
# 2 300 1000 50
learner$predict_newdata(new_data)
# <PredictionClassif> for 2 observations:
# row_ids truth response
# 1 <NA> Overt
# 2 <NA> Overt
# 保存模型
saveRDS(learner, 'diabetes_knn.rds')
model = readRDS('diabetes_knn.rds')
model$predict_newdata(new_data)
|

