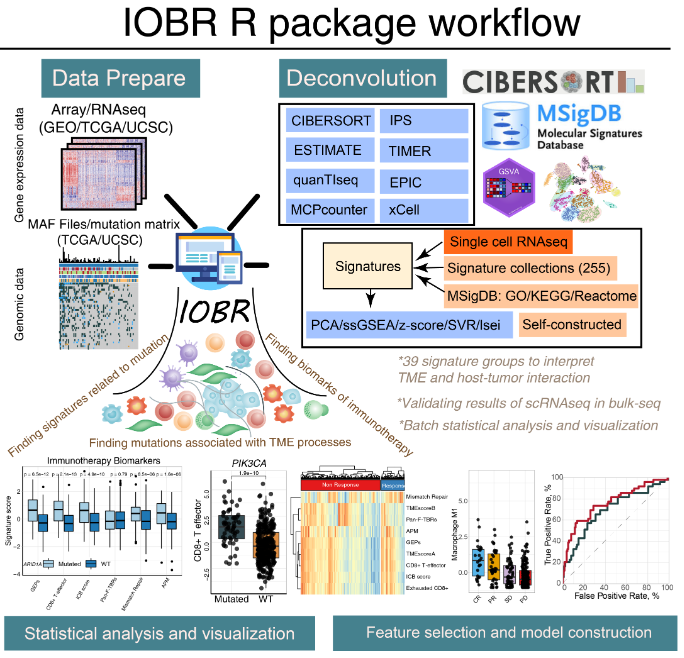
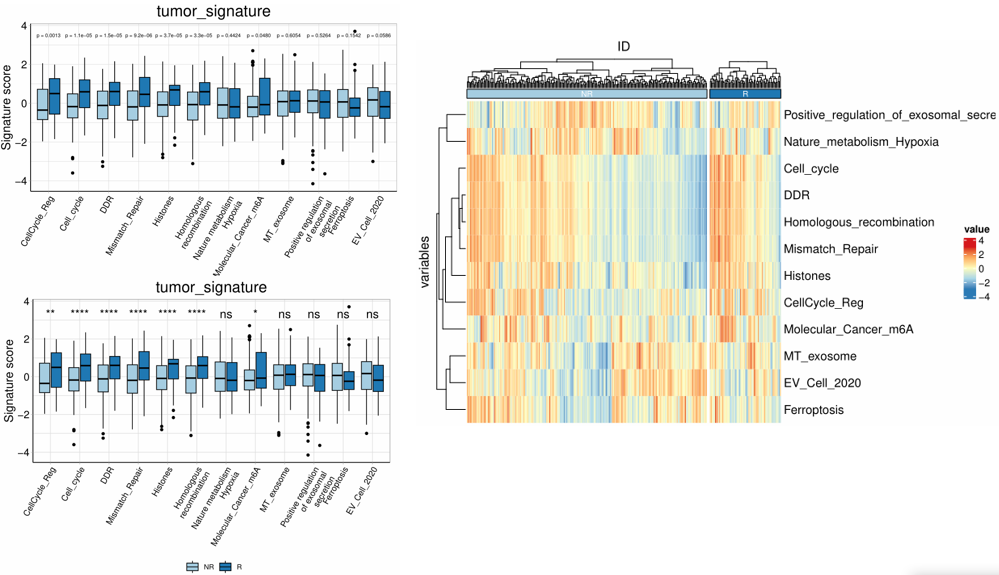
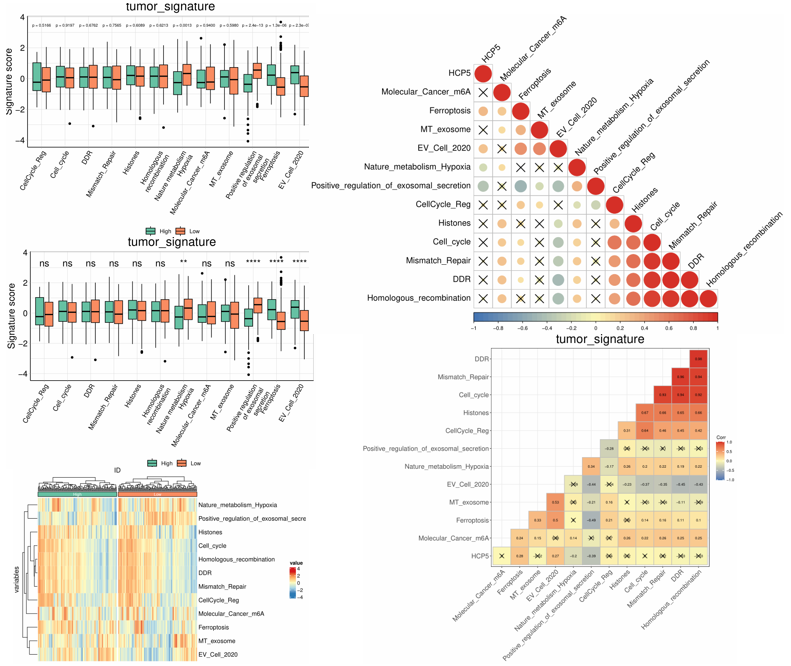
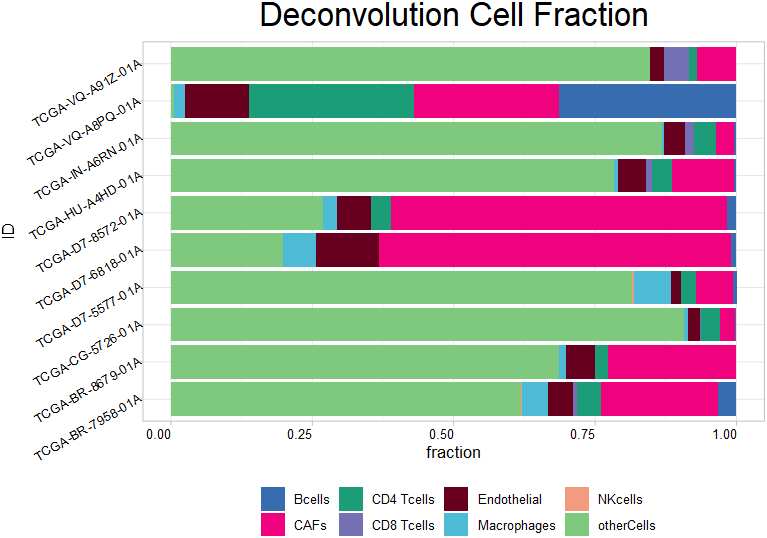
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
|
# options("repos"= c(CRAN="https://mirrors.tuna.tsinghua.edu.cn/CRAN/"))
# options(BioC_mirror="http://mirrors.tuna.tsinghua.edu.cn/bioconductor/")
if (!requireNamespace("BiocManager", quietly = TRUE)) install.packages("BiocManager")
depens<-c('tibble', 'survival', 'survminer', 'sva', 'limma', "DESeq2","devtools",
'limSolve', 'GSVA', 'e1071', 'preprocessCore', 'ggplot2', "biomaRt",
'ggpubr', "devtools", "tidyHeatmap", "caret", "glmnet", "ppcor","timeROC","pracma")
for(i in 1:length(depens)){
depen<-depens[i]
if (!requireNamespace(depen, quietly = TRUE)) BiocManager::install(depen)
}
if (!requireNamespace("remotes", quietly = TRUE)) install("remotes")
if (!requireNamespace("EPIC", quietly = TRUE))
remotes::install_github("GfellerLab/EPIC", build_vignettes=TRUE)
if (!requireNamespace("MCPcounter", quietly = TRUE))
remotes::install_github("ebecht/MCPcounter",ref="master", subdir="Source")
if (!requireNamespace("estimate", quietly = TRUE)){
rforge <- "http://r-forge.r-project.org"
install.packages("estimate", repos=rforge, dependencies=TRUE)
}
if (!requireNamespace("IOBR", quietly = TRUE))
remotes::install_github("IOBR/IOBR",ref="master")
library(IOBR)
|