1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
|
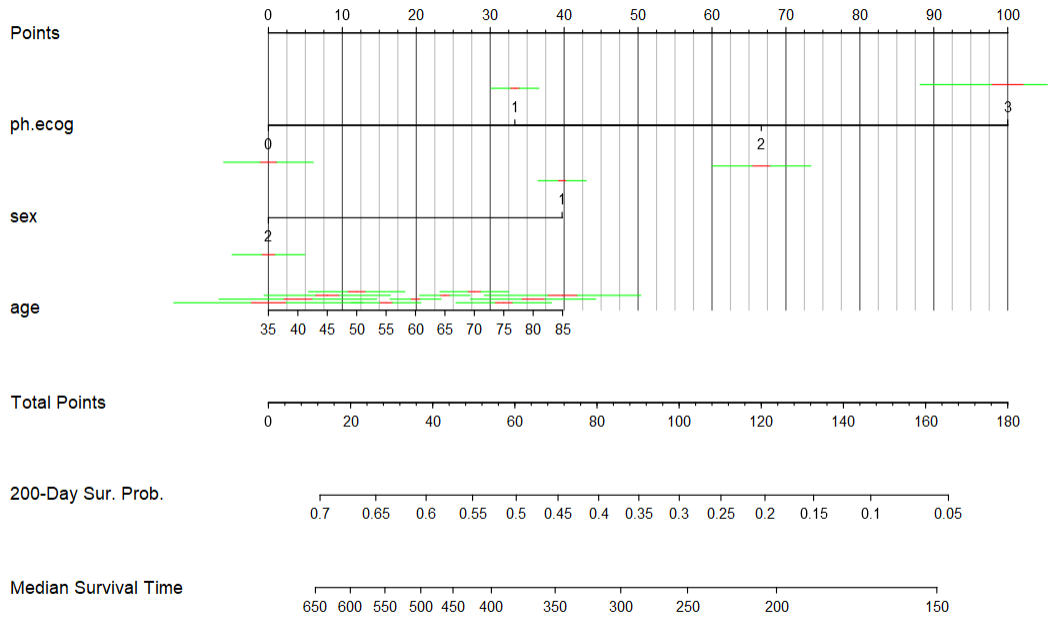
mod.cox.1 <- cph(Surv(time,status) ~ ph.ecog+sex+age,lung, x=T,y=T,surv = T, time.inc = 365)
cal1 <- calibrate(mod.cox.1, cmethod='KM', method="boot", u=365, m=60, B=1000)
mod.cox.2 <- cph(Surv(time,status) ~ ph.ecog+sex+age,lung, x=T,y=T,surv = T, time.inc = 365*2)
cal2 <- calibrate(mod.cox.2, cmethod='KM', method="boot", u=365*2, m=60, B=1000)
par(mar=c(7,5,1,1),cex = 0.75)
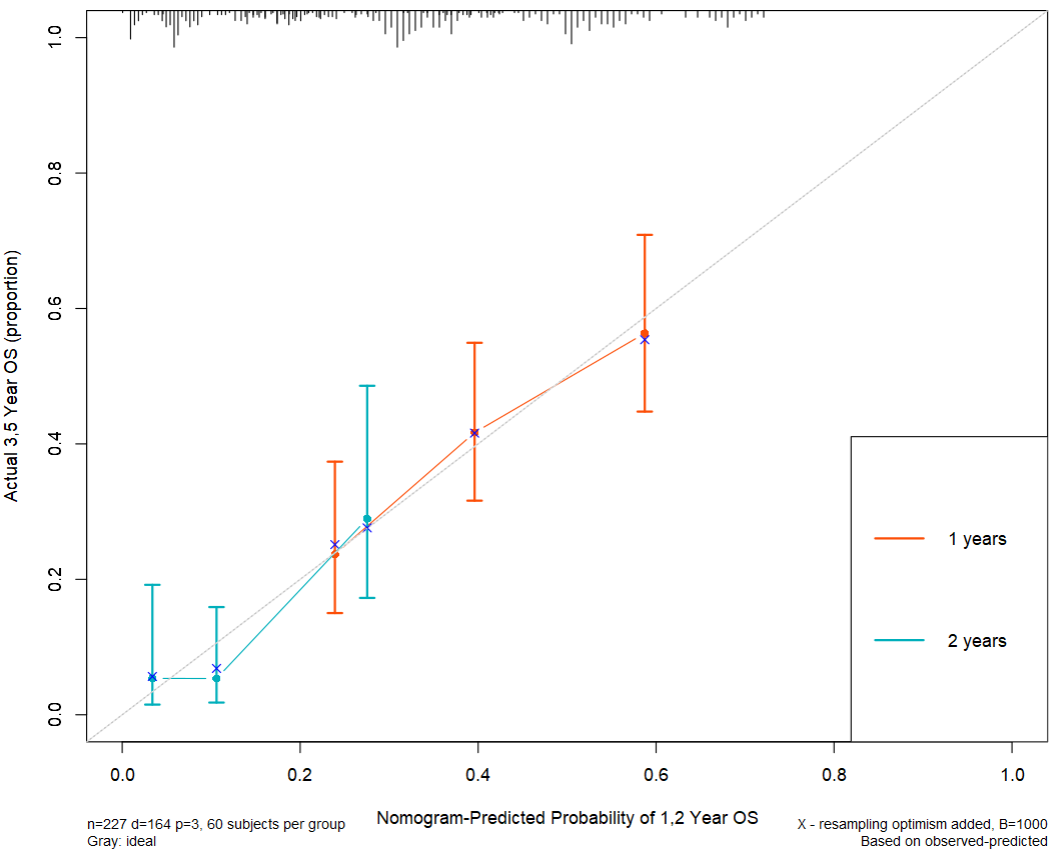
plot(cal1,lwd=2,lty=1,
errbar.col="#FC4E07", #线上面的竖线
xlim=c(0,1),ylim=c(0,1),
xlab="Nomogram-Predicted Probability of 1,2 Year OS",
ylab="Actual 1,1 Year OS (proportion)",
col="#FC4E07")
plot(cal2, add=T, conf.int=T,
subtitles = F, cex.subtitles=0.8,
lwd=2, lty=1, errbar.col="#00AFBB", col="#00AFBB")
#加上图例
legend("bottomright", legend=c("1 years", "2 years"),
col=c("#FC4E07","#00AFBB"), lwd=2)
#调整对角线
abline(0,1,lty=3,lwd=1,col="grey")
|