1、TCGAbiolinks下载数据
- 使用TCGAbiolinks下载特定肿瘤类型的SNV数据
- https://bioconductor.org/packages/release/bioc/vignettes/TCGAbiolinks/inst/doc/mutation.html
|
|
2、maftools可视化
- https://bioconductor.org/packages/release/bioc/vignettes/maftools/inst/doc/maftools.html
- https://www.jieandze1314.com/post/cnposts/237/
|
|
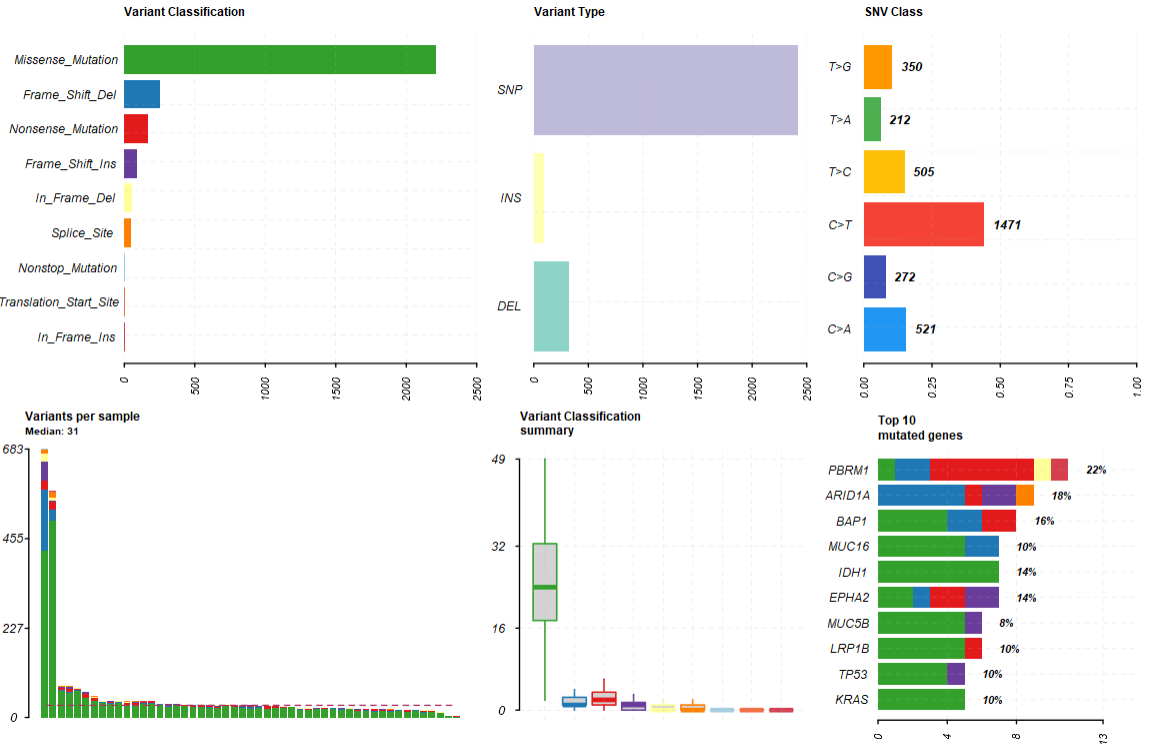
2.1概括图
|
|
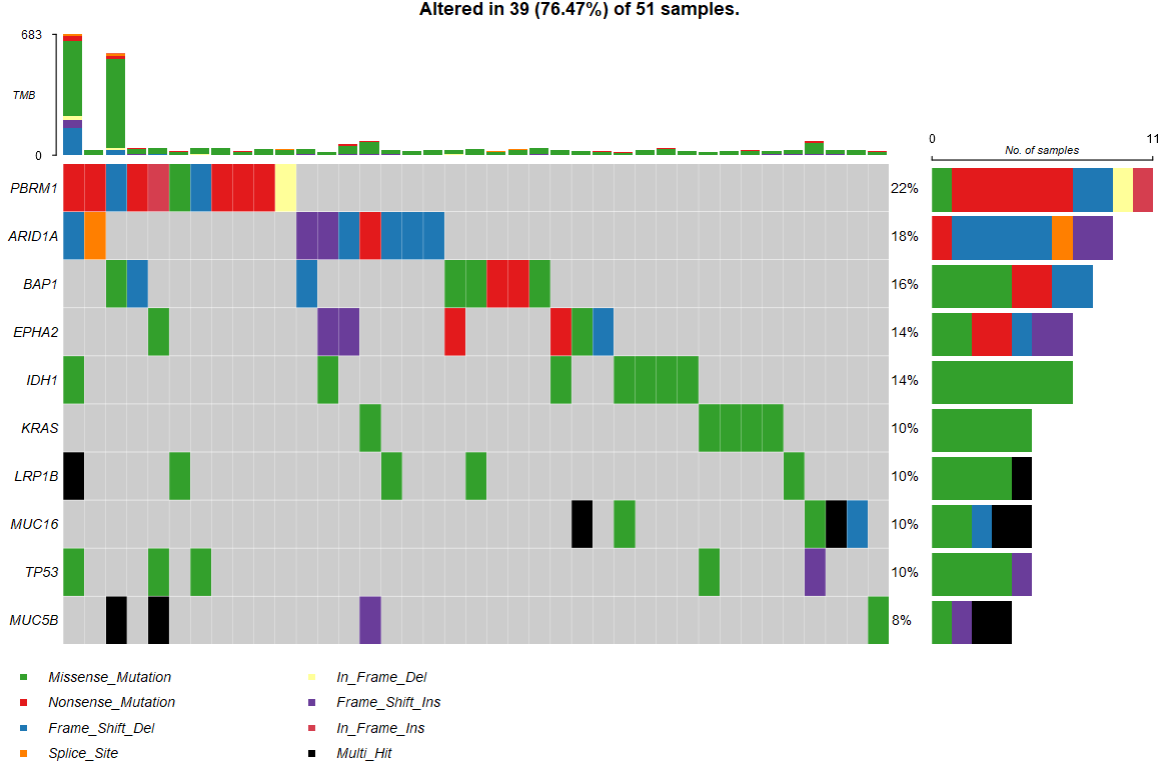
2.2 基因瀑布图
|
|
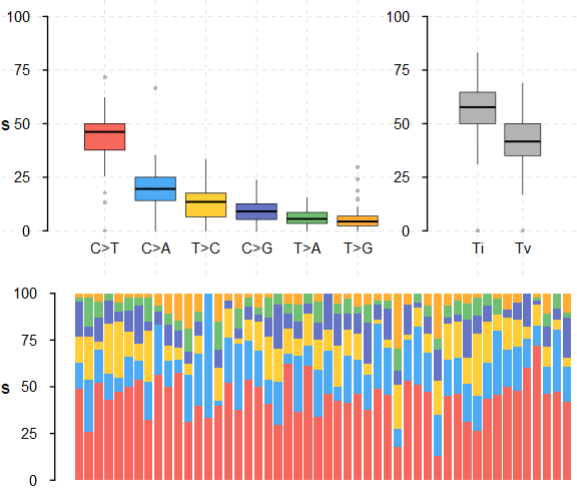
2.3 转换颠倒统计
|
|
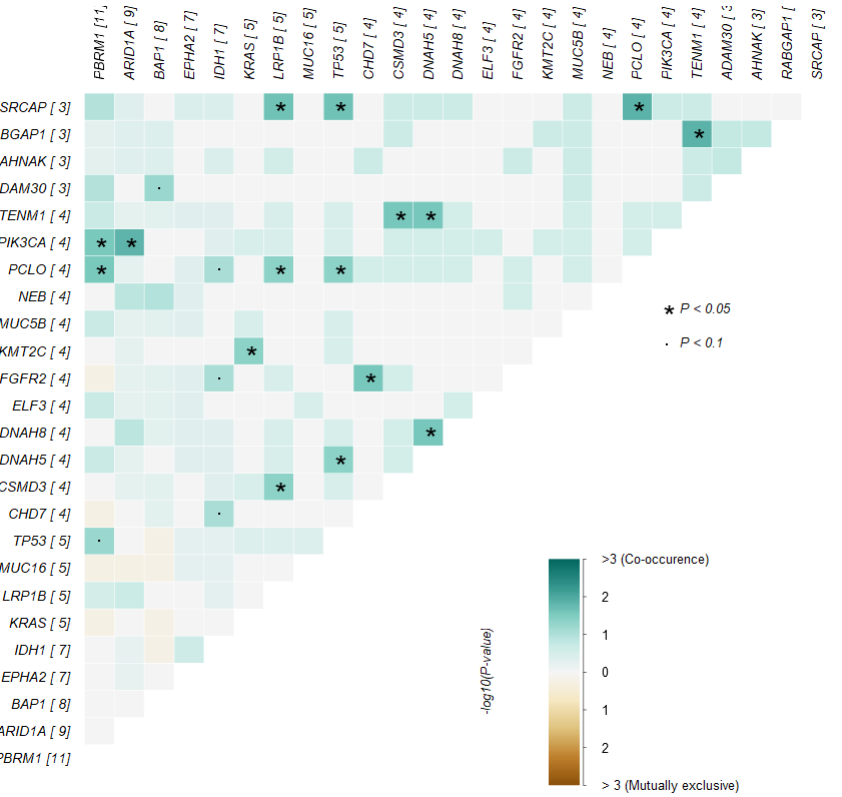
2.4 基因对突变统计
|
|
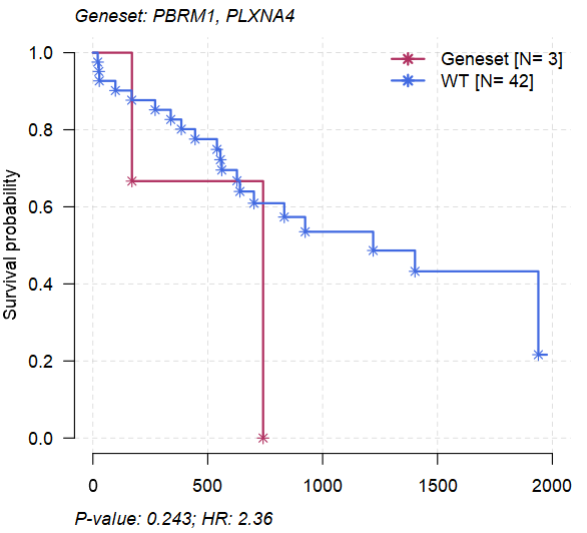
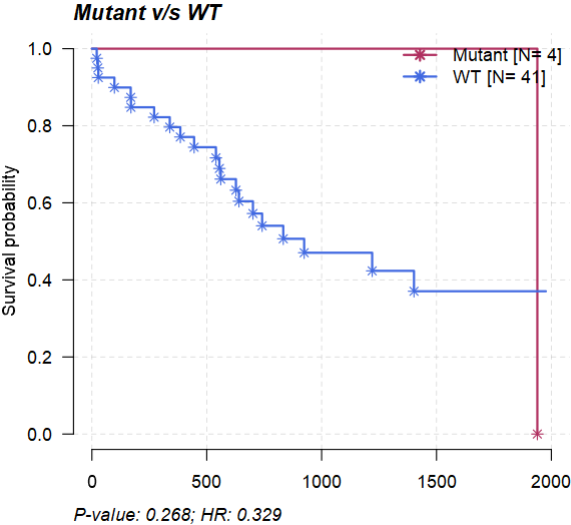
2.5 生存分析
- 根据特定基因是否突变将病人分成WT与Mutant两组
|
|
2.6 基因对的生存相关性
|
|