1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
|
pdf('tmp1.pdf',height = 10,width = 12,onefile = F)
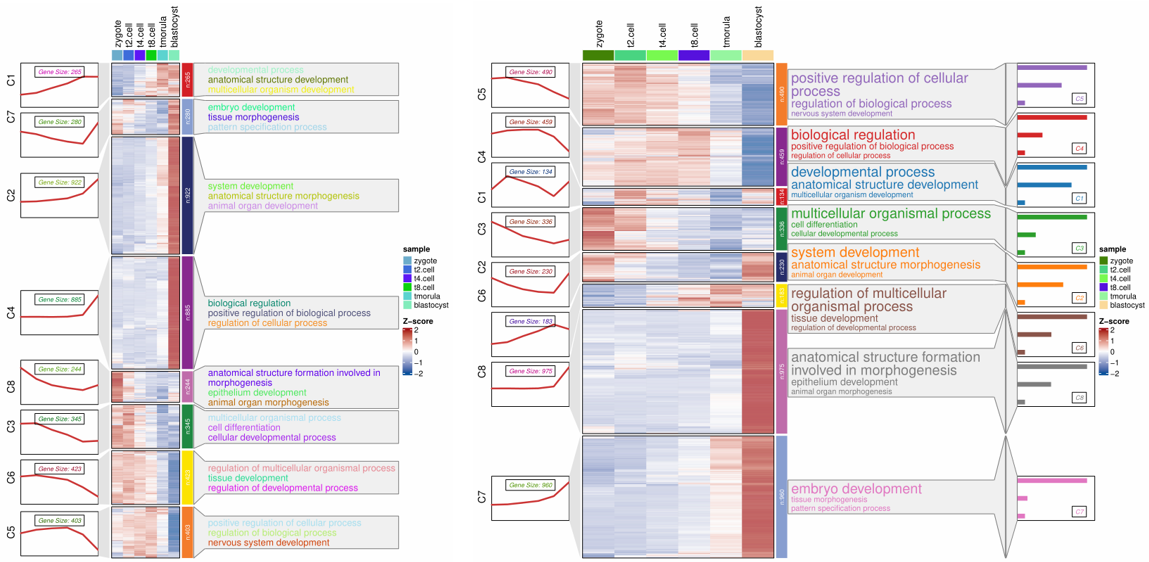
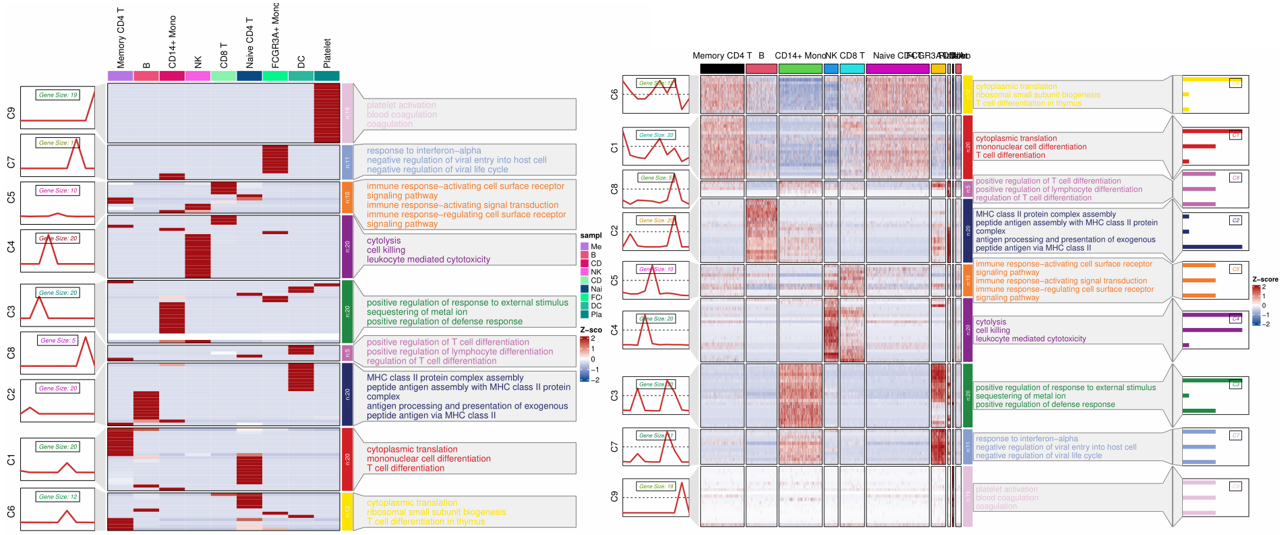
visCluster(object = st.data1,
plot.type = "both",
show_row_dend = F,
markGenes.side = "left",
annoTerm.data = enrich,
line.side = "left",
go.col = rep(jjAnno::useMyCol("stallion",n = 9),each = 3))
dev.off()
pdf('tmp2.pdf',height = 10,width = 14,onefile = F)
visCluster(object = st.data3,
plot.type = "both",
show_row_dend = F,
markGenes.side = "left",
annoTerm.data = enrich,
line.side = "left",
go.col = rep(jjAnno::useMyCol("stallion",n = 9),each = 3),
add.bar = T,
annoTerm.mside = "left",
show_column_names = F)
dev.off()
|