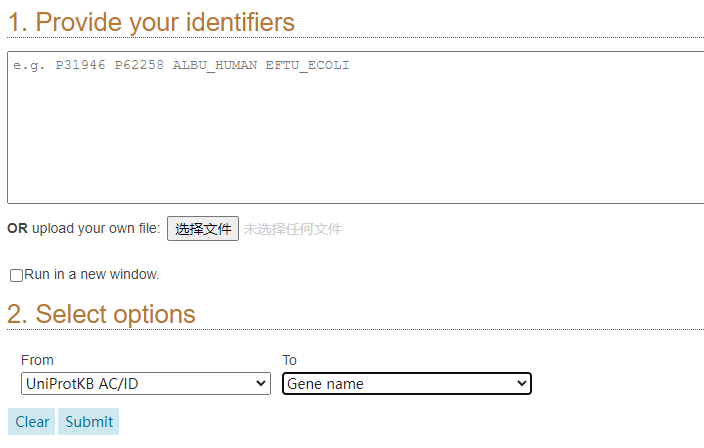
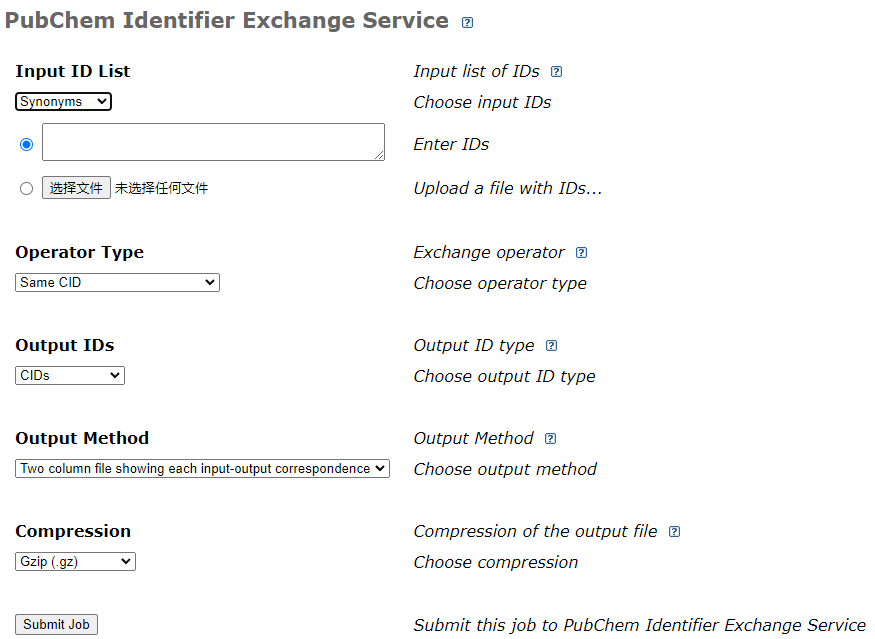
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
|
c = pcp.Compound.from_cid(5090)
type(c)
# pubchempy.Compound
##举例3个属性值
c.molecular_formula
# 'C17H14O4S'
c.canonical_smiles
# 'CS(=O)(=O)C1=CC=C(C=C1)C2=C(C(=O)OC2)C3=CC=CC=C3'
c.synonyms[:3]
# ['rofecoxib', '162011-90-7', 'Vioxx']
## 转换成表格,显示全部属性
c_df = pcp.compounds_to_frame(c)
c_df.columns
# Index(['atom_stereo_count', 'atoms', 'bond_stereo_count', 'bonds',
# 'cactvs_fingerprint', 'canonical_smiles', 'charge', 'complexity',
# 'conformer_id_3d', 'conformer_rmsd_3d', 'coordinate_type',
# 'covalent_unit_count', 'defined_atom_stereo_count',
# 'defined_bond_stereo_count', 'effective_rotor_count_3d', 'elements',
# 'exact_mass', 'feature_selfoverlap_3d', 'fingerprint',
# 'h_bond_acceptor_count', 'h_bond_donor_count', 'heavy_atom_count',
# 'inchi', 'inchikey', 'isomeric_smiles', 'isotope_atom_count',
# 'iupac_name', 'mmff94_energy_3d', 'mmff94_partial_charges_3d',
# 'molecular_formula', 'molecular_weight', 'monoisotopic_mass',
# 'multipoles_3d', 'pharmacophore_features_3d', 'record',
# 'rotatable_bond_count', 'shape_fingerprint_3d', 'shape_selfoverlap_3d',
# 'tpsa', 'undefined_atom_stereo_count', 'undefined_bond_stereo_count',
# 'volume_3d', 'xlogp'],
# dtype='object')
|