1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
|
library(TCGAbiolinks)
library(SummarizedExperiment)
library(tidyverse)
query <- GDCquery(project = "TCGA-CHOL",
legacy = FALSE,
data.category = "Transcriptome Profiling",
data.type = "Gene Expression Quantification")
GDCdownload(query, files.per.chunk = 10)
data = GDCprepare(query, save = F)
rownames(data) = rowData(data)$gene_name
data = data[rowData(data)$gene_type=="protein_coding",]
count = assay(data, "unstranded")
count = count[!duplicated(rownames(count)),]
dim(count)
# [1] 19938 44
count[1:4,1:4]
# TCGA-W5-AA2X-01A-11R-A41I-07 TCGA-W5-AA2X-11A-11R-A41I-07
# TSPAN6 3310 2322
# TNMD 0 0
# DPM1 1881 716
# SCYL3 966 315
# TCGA-W5-AA33-01A-11R-A41I-07 TCGA-ZH-A8Y2-01A-11R-A41I-07
# TSPAN6 11312 6768
# TNMD 1 0
# DPM1 968 1273
# SCYL3 1296 774
meta = colData(data)[,c("barcode","sample", "patient","shortLetterCode")] %>%
as.data.frame()
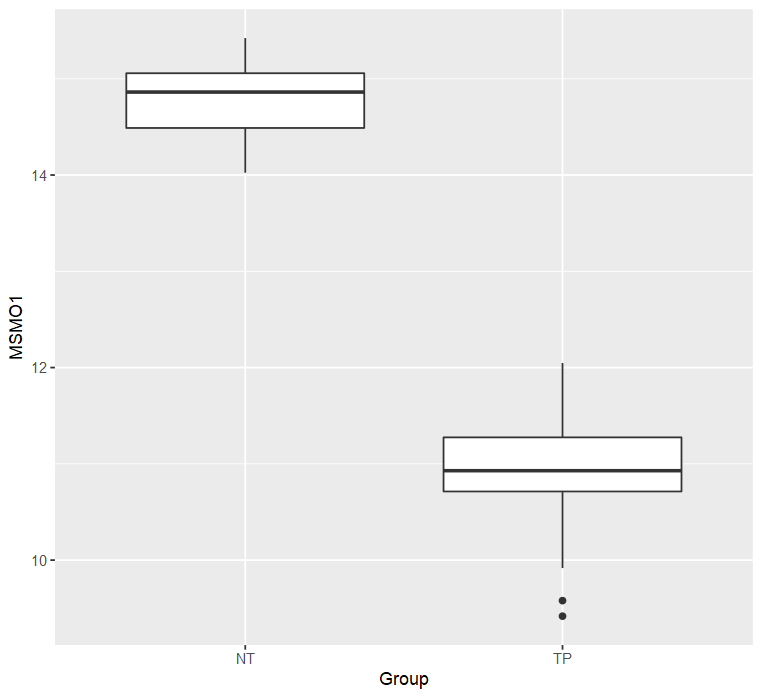
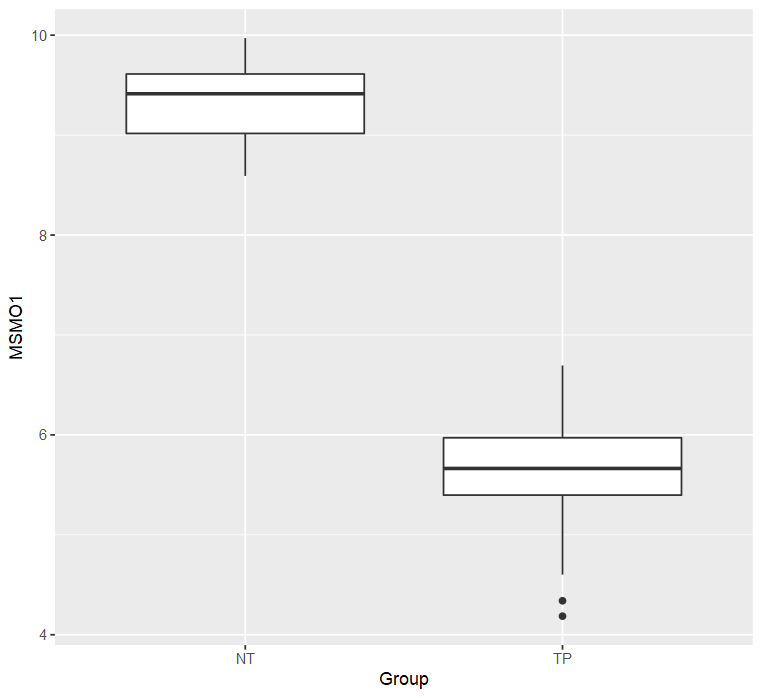
meta$Group = meta$shortLetterCode # NT: Solid Tissue Normal; TP: Primary Tumor
head(meta[,"Group",drop=F])
# Group
# TCGA-W5-AA2X-01A-11R-A41I-07 TP
# TCGA-W5-AA2X-11A-11R-A41I-07 NT
# TCGA-W5-AA33-01A-11R-A41I-07 TP
# TCGA-ZH-A8Y2-01A-11R-A41I-07 TP
# TCGA-ZH-A8Y6-01A-11R-A41I-07 TP
# TCGA-W5-AA2Z-01A-11R-A41I-07 TP
table(meta$Group)
# NT TP
# 9 35
identical(colnames(count), rownames(meta))
# [1] TRUE
## 8对配对样本
meta_pair = meta[TCGAquery_MatchedCoupledSampleTypes(meta$barcode, c("NT","TP")), ]
# TCGA-W5-AA2I TCGA-W5-AA2Q TCGA-W5-AA2U TCGA-W5-AA2X TCGA-W5-AA30 TCGA-W5-AA31 TCGA-W5-AA34 TCGA-ZU-A8S4
# 2 2 2 2 2 2 2 2
count_pair = count[, meta_pair$barcode]
identical(meta_pair$barcode, colnames(count_pair))
#[1] TRUE
|