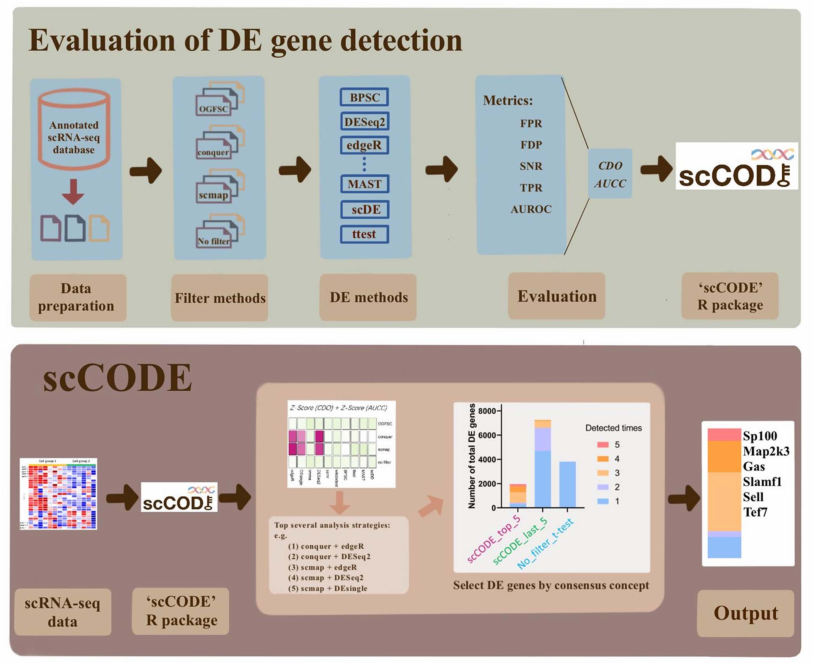
scCODE( single-cell consensus optimization of differentially expressed gene detection)是由复旦大学附属金山医院邹欣等人开发的R包工具,于2022年12月发表于Briefing in Bioinformatics;该工具对多种差异基因分析策略进行了集成、整合,用于鉴定鲁棒性的单细胞差异基因。用法比较简单,简单记录如下。
- Paper:https://academic.oup.com/bib/article-abstract/23/5/bbac180/6590434?redirectedFrom=fulltext
- Github:https://github.com/XZouProjects/scCODE
1、安装R包
|
|
2、差异基因分析
- (1)准备两组单细胞样本的count表达矩阵
|
|
- (2)差异分析
-
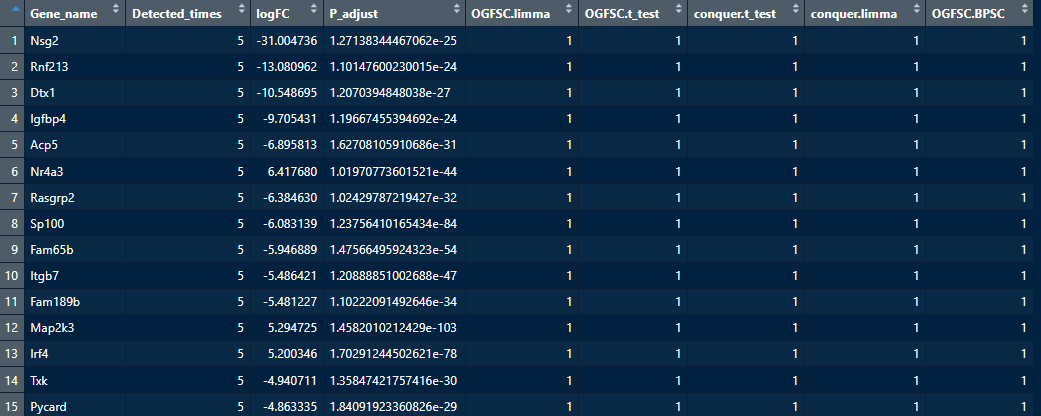
默认light模式下,使用5种策略进行分析;再统计每种策略的判断结果。
-
如果一个基因的5种结果均判断为显著差异基因,则相对更可靠。
-
在linux端使用时,出现类似
OpenBLAS blas_thread_init: pthread_create failed for thread 60 of 128: Resource temporarily unavailable报错,经查在shell命令行设置如下参数可正常使用。1 2 3export OPENBLAS_NUM_THREADS=2 export GOTO_NUM_THREADS=2 export OMP_NUM_THREADS=2
-
|
|